4NRQ
 
 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
4NRP
 
 | |
4O7X
 
 | |
4NRM
 
 | |
4NRO
 
 | |
2C9X
 
 | | Sulfite dehydrogenase from Starkeya Novella Y236F mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U, Feng, C, Honeychurch, M.J, Bernhardt, P, Tollin, G, Enemark, J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Evidence for the Importance of Tyr236 for the Integrity of the Mo Active Site in a Bacterial Sulfite Dehydrogenase.
Biochemistry, 45, 2006
|
|
3LNW
 
 | |
3A9U
 
 | |
3A9V
 
 | |
3NI2
 
 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | Authors: | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
4N0N
 
 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4N0O
 
 | | Complex structure of Arterivirus nonstructural protein 10 (helicase) with DNA | | Descriptor: | CALCIUM ION, DNA, Replicase polyprotein 1ab, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
6LRM
 
 | | Crystal structure of PDE4D catalytic domain in complex with arctigenin | | Descriptor: | 1,2-ETHANEDIOL, Arctigenin, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-01-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of phosphodiesterase-4 as the therapeutic target of arctigenin in alleviating psoriatic skin inflammation.
J Adv Res, 33, 2021
|
|
8P2L
 
 | |
8P2M
 
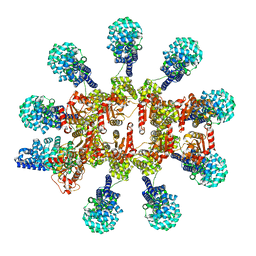 | | C. elegans TIR-1 protein. | | Descriptor: | NAD(+) hydrolase tir-1 | | Authors: | Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans.
Cell Rep, 42, 2023
|
|
6E20
 
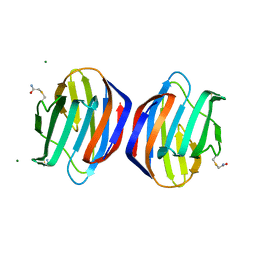 | | Crystal structure of the Dario rerio galectin-1-L2 | | Descriptor: | Galectin, MAGNESIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Ghosh, A, Bianchet, M.A. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the zebrafish galectin-1-L2 and model of its interaction with the infectious hematopoietic necrosis virus (IHNV) envelope glycoprotein.
Glycobiology, 29, 2019
|
|
6MX5
 
 | |
2CA3
 
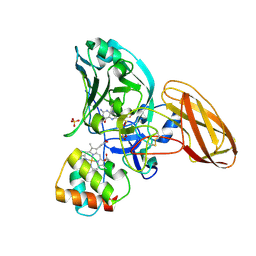 | | Sulfite dehydrogenase from Starkeya Novella r55m mutant | | Descriptor: | (MOLYBDOPTERIN-S,S)-OXO-MOLYBDENUM, HEME C, SULFATE ION, ... | | Authors: | Bailey, S, Kappler, U, Feng, C, Honeychurch, M.J, Bernhardt, P.V, Tollin, G, Enemark, J.H. | | Deposit date: | 2005-12-16 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for enzymatic sulfite oxidation: how three conserved active site residues shape enzyme activity.
J.Biol.Chem., 284, 2009
|
|
6DXI
 
 | |
6IMB
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMI
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6-ethoxy-7-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6INK
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMD
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X, Su, H, Xu, Y. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IND
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-6,7-dimethoxy-1-[2-(6-methyl-1H-indol-3-yl)ethyl]-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IM6
 
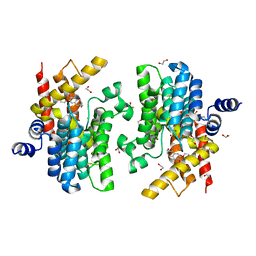 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 7-ethoxy-6-methoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
