5ZBR
 
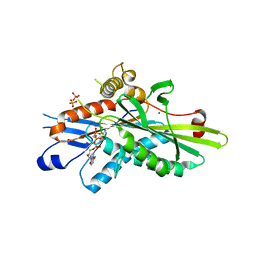 | | Crystal Structure of Kinesin-3 KIF13B motor domain in AMPPNP form | | Descriptor: | Kinesin family member 13B, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Delineation of the Neck Linker of Kinesin-3 for Processive Movement.
J. Mol. Biol., 430, 2018
|
|
7WRG
 
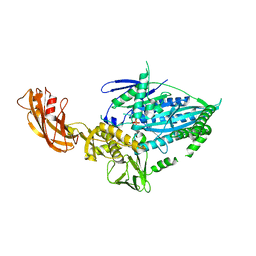 | | Crystal structure of full-length kinesin-3 KLP-6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein, MAGNESIUM ION | | Authors: | Wang, W.J, Ren, J.Q, Song, W.Y, Feng, W. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | The architecture of kinesin-3 KLP-6 reveals a multilevel-lockdown mechanism for autoinhibition.
Nat Commun, 13, 2022
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7WEJ
 
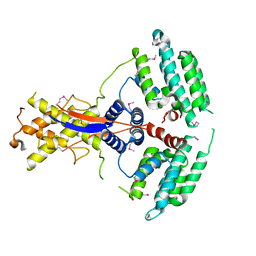 | | Crystal structure of the mouse Wdr47 NTD. | | Descriptor: | WD repeat-containing protein 47 | | Authors: | Ren, J.Q, Li, D, Feng, W. | | Deposit date: | 2021-12-23 | | Release date: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Intertwined Wdr47-NTD dimer recognizes a basic-helical motif in Camsap proteins for proper central-pair microtubule formation.
Cell Rep, 41, 2022
|
|
7WEK
 
 | |
7XKC
 
 | | Crystal structure of Daucus Carrot hypoglycemic peptide (DCHP) | | Descriptor: | DCHP, SULFATE ION | | Authors: | Guo, T, Ren, J.Q, Wang, L, Shi, Y.W, Feng, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Characterization of a thermostable, protease-tolerant inhibitor of alpha-glycosidase from carrot: A potential oral additive for treatment of diabetes.
Int.J.Biol.Macromol., 209, 2022
|
|
7D8V
 
 | | Crystal Structure of A Kinesin-3 KIF13B mutant-T192Y | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin family member 13B, ... | | Authors: | Ren, J.Q, Feng, W. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motor domain-mediated autoinhibition dictates axonal transport by the kinesin UNC-104/KIF1A.
Plos Genet., 17, 2021
|
|
7YQH
 
 | | Cryo-EM structure of 8-subunit Smc5/6 | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 3, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2022-08-07 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of 8-subunit Smc5/6
To Be Published
|
|
7YMD
 
 | | Cryo-EM structure of Nse1/3/4 | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | Authors: | Qian, L, Jun, Z, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.176 Å) | | Cite: | Cryo-EM structure of Nse1/3/4
To Be Published
|
|
8IOD
 
 | | Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8INR
 
 | | Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8IOC
 
 | | Cryo-EM structure of the gamma-MSH-bound human melanocortin receptor 3 (MC3R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
4N0N
 
 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4N0O
 
 | | Complex structure of Arterivirus nonstructural protein 10 (helicase) with DNA | | Descriptor: | CALCIUM ION, DNA, Replicase polyprotein 1ab, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
5ET0
 
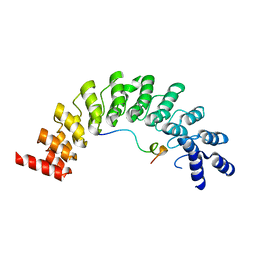 | | Crystal structure of Myo3b-ARB2 in complex with Espin1-AR | | Descriptor: | Espin, Myosin-IIIb | | Authors: | Liu, H, Li, J, Liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
5ET1
 
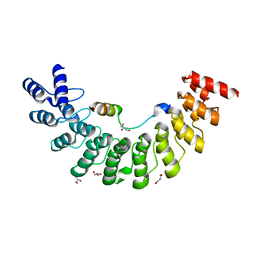 | | Crystal structure of Myo3b-ARB1 in complex with Espin1-AR | | Descriptor: | Espin, GLYCEROL, Myosin-IIIb | | Authors: | Liu, H, Li, J, liu, W, Zhang, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Myosin III-mediated cross-linking and stimulation of actin bundling activity of Espin
Elife, 5, 2016
|
|
2KXS
 
 | | ZO1 ZU5 domain in complex with GRINL1A peptide | | Descriptor: | Tight junction protein ZO-1,Myocardial zonula adherens protein | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-30 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Cdc42-dependent formation of the ZO-1/MRCKb complex at the leading edge controls cell migration
Embo J., 30, 2011
|
|
2KXR
 
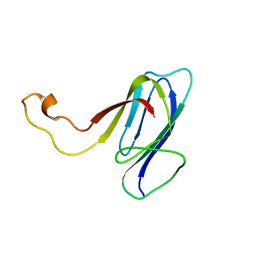 | | ZO1 ZU5 domain MC/AA mutation | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cdc42-dependent formation of the ZO-1/MRCKb complex at the leading edge controls cell migration
Embo J., 30, 2011
|
|
5AZF
 
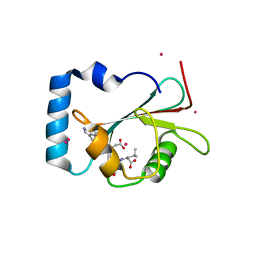 | | Crystal structure of LGG-1 complexed with a WEEL peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, SULFATE ION, ... | | Authors: | Watanabe, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AZG
 
 | | Crystal structure of LGG-1 complexed with a UNC-51 peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, Serine/threonine-protein kinase unc-51 | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AZH
 
 | | Crystal structure of LGG-2 fused with an EEEWEEL peptide | | Descriptor: | EEEWEEL peptide,Protein lgg-2, MAGNESIUM ION | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
3WDO
 
 | | Structure of E. coli YajR transporter | | Descriptor: | MFS Transporter | | Authors: | Jiang, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4Q2M
 
 | |
4Q2L
 
 | |
