3BN9
 
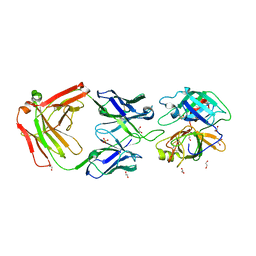 | | Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 | | Descriptor: | 1,2-ETHANEDIOL, E2 Fab Heavy Chain, E2 Fab Light Chain, ... | | Authors: | Farady, C.J, Schneider, E.L, Egea, P.F, Goetz, D.H, Craik, C.S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition
J.Mol.Biol., 380, 2008
|
|
3SO3
 
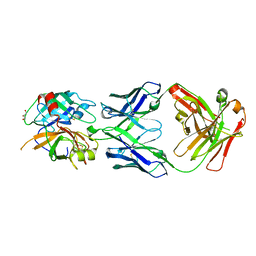 | | Structures of Fab-Protease Complexes Reveal a Highly Specific Non-Canonical Mechanism of Inhibition. | | Descriptor: | A11 FAB heavy chain, A11 FAB light chain, GLYCEROL, ... | | Authors: | Schneider, E.L, Farady, C.J, Egea, P.F, Goetz, D.H, Baharuddin, A, Craik, C.S. | | Deposit date: | 2011-06-29 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A reverse binding motif that contributes to specific protease inhibition by antibodies.
J.Mol.Biol., 415, 2012
|
|
6FFS
 
 | | Structure-based design and synthesis of macrocyclic human rhinovirus 3C protease inhibitors | | Descriptor: | 3C Protease, SULFATE ION, ~{N}-[(2~{S},5~{S},14~{S})-2-[(4-fluorophenyl)methyl]-5-(hydroxymethyl)-9-methyl-3,8,15-tris(oxidanylidene)-1,4,9-triazacyclopentadec-14-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wiesmann, C, Farady, C. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based design and synthesis of macrocyclic human rhinovirus 3C protease inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6FFN
 
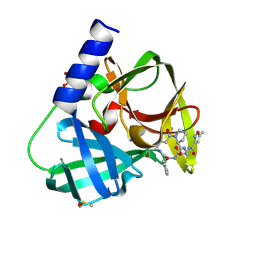 | |
2OZ2
 
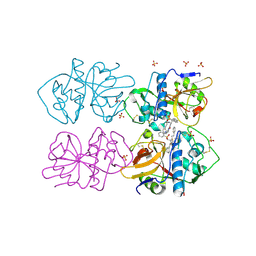 | |
2P7U
 
 | |
7ALV
 
 | | Crystal Structure of NLRP3 NACHT domain in complex with a potent inhibitor | | Descriptor: | 1-[4-chloranyl-2,6-di(propan-2-yl)phenyl]-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | Crystal Structure of NLRP3 NACHT Domain With an Inhibitor Defines Mechanism of Inflammasome Inhibition.
J.Mol.Biol., 433, 2021
|
|
8RI2
 
 | | Crystal structure of NLRP3 in complex with inhibitor NP3-562 | | Descriptor: | 2-[4-chloranyl-9-oxidanylidene-12-(2-oxidanylpropan-2-yl)-5-thia-1,10,11-triazatricyclo[6.4.0.0^{2,6}]dodeca-2(6),3,7,11-tetraen-10-yl]-~{N}-[(3~{R})-1-methylpiperidin-3-yl]ethanamide, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Dekker, C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent, Orally Bioavailable, Tricyclic NLRP3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
3BWK
 
 | | Crystal Structure of Falcipain-3 with Its inhibitor, K11017 | | Descriptor: | Cysteine protease falcipain-3, N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide, SULFATE ION | | Authors: | Kerr, I, Lee, J.H, Brinen, L.S. | | Deposit date: | 2008-01-09 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Vinyl sulfones as antiparasitic agents and a structural basis for drug design.
J.Biol.Chem., 284, 2009
|
|
3NPS
 
 | |
