4YEY
 
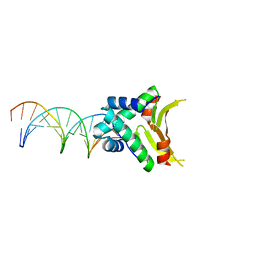 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
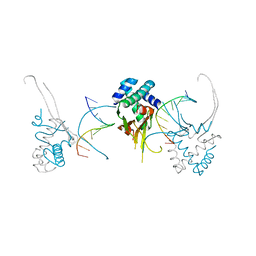 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YC8
 
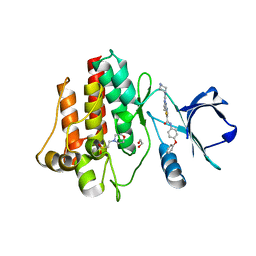 | | C-Helix-Out Binding of Dasatinib Analog to c-Abl Kinase | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(4-phenoxyphenyl)-1,3-thiazole-5-carboxamide, ... | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.J, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-19 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
4YF0
 
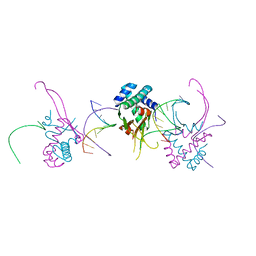 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
1IU5
 
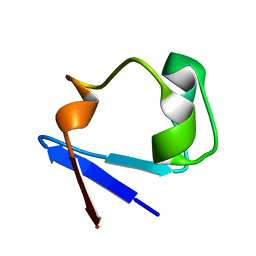 | | X-ray Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IU6
 
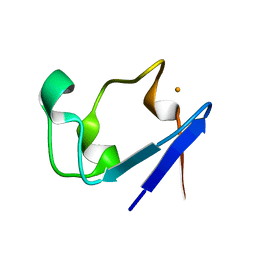 | | Neutron Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4YBJ
 
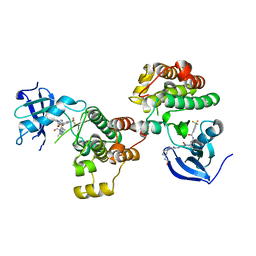 | | Type II Dasatinib Analog Crystallized with c-Src Kinase | | Descriptor: | 2-({6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-methylpyrimidin-4-yl}amino)-N-(3-{[3-(trifluoromethyl)benzoyl]amino}phenyl)-1,3-thiazole-5-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Kwarcinski, F.E, Brandvold, K.B, Johnson, T.K, Phadke, S, Meagher, J.L, Seeliger, M.A, Stuckey, J.A, Soellner, M.B. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Acs Chem.Biol., 11, 2016
|
|
1KBJ
 
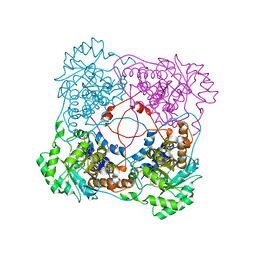 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: comparison with the Intact Wild-type Enzyme | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
1KBI
 
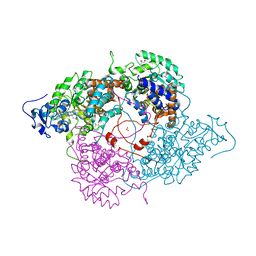 | | Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME B2, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.-W, Welsh, F.E, Chapman, S.K, Reid, G.A, Mathews, F.S. | | Deposit date: | 2001-11-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic study of the recombinant flavin-binding domain of Baker's yeast flavocytochrome b(2): comparison with the intact wild-type enzyme.
Biochemistry, 41, 2002
|
|
5AKO
 
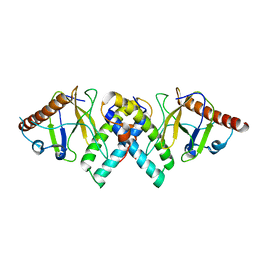 | |
1LE5
 
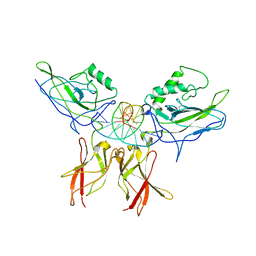 | | Crystal structure of a NF-kB heterodimer bound to an IFNb-kB | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*T)-3', Nuclear factor NF-kappa-B p50 subunit, ... | | Authors: | Berkowitz, B, Huang, D.B, Chen-Park, F.E, Sigler, P.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The X-ray crystal structure of the
NF-kB p50/p65 heterodimer bound
to the Interferon beta-kB site
J.Biol.Chem., 277, 2002
|
|
1LE9
 
 | | Crystal structure of a NF-kB heterodimer bound to the Ig/HIV-kB siti | | Descriptor: | 5'-D(*AP*AP*GP*GP*AP*AP*AP*GP*TP*CP*CP*C)-3', 5'-D(*TP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*T)-3', NUCLEAR FACTOR NF-KAPPA-B P50 SUBUNIT, ... | | Authors: | Benjamin, B, Huang, D.B, Chen-Park, F.E, Sigler, P.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The x-ray crystal structure of the NF-kappa B p50.p65 heterodimer bound to the interferon beta -kappa B site.
J.Biol.Chem., 277, 2002
|
|
3SD3
 
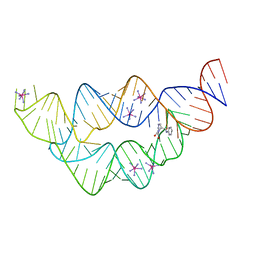 | | The structure of the tetrahydrofolate riboswitch containing a U25C mutation | | Descriptor: | IRIDIUM HEXAMMINE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Tetrahydrofolate riboswitch | | Authors: | Reyes, F.E, Trausch, J.J, Ceres, P, Batey, R.T. | | Deposit date: | 2011-06-08 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer.
Structure, 19, 2011
|
|
3ZHN
 
 | | Crystal structure of the T6SS lipoprotein TssJ1 from Pseudomonas aeruginosa | | Descriptor: | IODIDE ION, PA_0080 | | Authors: | Robb, C.S, Assmus, M, Nano, F.E, Boraston, A.B. | | Deposit date: | 2012-12-22 | | Release date: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the T6Ss Lipoprotein Tssj1 from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1NDT
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (NITRITE REDUCTASE) | | Authors: | Dodd, F.E, Vanbeeumen, J, Eady, R.R, Hasnain, S.S. | | Deposit date: | 1998-10-28 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner.
J.Mol.Biol., 282, 1998
|
|
1NDR
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1NDS
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A SUBSTRATE BOUND BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1OE1
 
 | |
1OE3
 
 | | Atomic resolution structure of 'Half Apo' NiR | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL | | Authors: | Ellis, M.J, Dodd, F.E, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2003-03-18 | | Release date: | 2004-07-21 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Native Copper Nitrite Reductase from Alcaligenes Xylosoxidans and the Active Site Mutant Asp92Glu
J.Mol.Biol., 328, 2003
|
|
1OE2
 
 | | Atomic Resolution Structure of D92E Mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL | | Authors: | Ellis, M.J, Dodd, F.E, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic Resolution Structures of Native Copper Nitrite Reductase from Alcaligenes Xylosoxidans and the Active Site Mutant Asp92Glu
J.Mol.Biol., 328, 2003
|
|
1PE3
 
 | |
1PY9
 
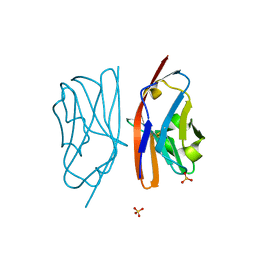 | | The crystal structure of an autoantigen in multiple sclerosis | | Descriptor: | Myelin-oligodendrocyte glycoprotein, SULFATE ION | | Authors: | Clements, C.S, Reid, H.H, Beddoe, T, Tynan, F.E, Perugini, M.A, Johns, T.G, Bernard, C.C, Rossjohn, J. | | Deposit date: | 2003-07-08 | | Release date: | 2003-09-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of myelin oligodendrocyte glycoprotein, a key autoantigen in multiple sclerosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1SIR
 
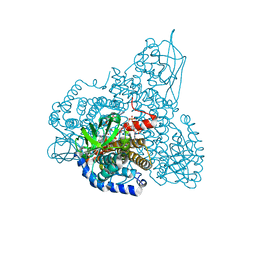 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, S-4-NITROBUTYRYL-COA | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S.L, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
1SIQ
 
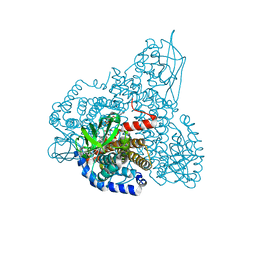 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
2ERX
 
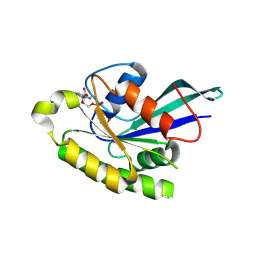 | | Crystal Structure of DiRas2 in Complex With GDP and Inorganic Phosphate | | Descriptor: | GTP-binding protein Di-Ras2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Papagrigoriou, E, Yang, X, Elkins, J, Niesen, F.E, Burgess, N, Salah, E, Fedorov, O, Ball, L.J, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of DiRas2
To be Published
|
|
