5AMT
 
 | | Intracellular growth locus protein E | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, IGLE | | Authors: | Robb, C.S, Nano, F.E, Boraston, A.B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Diffraction Analysis of Intracellular Growth Locus E (Igle) Protein from Francisella Tularensis Subsp. Novicida.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3BWA
 
 | | Crystal Structure of HLA B*3508 in complex with a HCMV 8-mer peptide from the pp65 protein | | Descriptor: | Beta-2-microglobulin, FPT peptide from 65 kDa lower matrix phosphoprotein, HLA class I histocompatibility antigen, ... | | Authors: | Wynn, K.K, Marland, Z, Cooper, L, Silins, S.L, Gras, S, Archbold, J.K, Tynan, F.E, Miles, J.J, McCluskey, J, Burrows, S.R, Rossjohn, J, Khanna, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection
Blood, 111, 2008
|
|
6CSF
 
 | | Crystal structure of sodium/alanine symporter AgcS with D-alanine bound | | Descriptor: | D-ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1NDS
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A SUBSTRATE BOUND BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1NDR
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1OE1
 
 | |
1OE3
 
 | | Atomic resolution structure of 'Half Apo' NiR | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL | | Authors: | Ellis, M.J, Dodd, F.E, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2003-03-18 | | Release date: | 2004-07-21 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Native Copper Nitrite Reductase from Alcaligenes Xylosoxidans and the Active Site Mutant Asp92Glu
J.Mol.Biol., 328, 2003
|
|
6G99
 
 | | Solution structure of FUS-ZnF bound to UGGUG | | Descriptor: | RNA (5'-R(*UP*GP*GP*UP*G)-3'), RNA-binding protein FUS, ZINC ION | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
8EEX
 
 | | Cas7-11 in complex with Csx29 | | Descriptor: | Cas7-11, Csx29, ZINC ION, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
8EEY
 
 | | Cas7-11 in complex with DR-mismatched target RNA, Csx29 and Csx30 | | Descriptor: | Cas7-11, Csx29, Csx30, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
6GBM
 
 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
1I17
 
 | | NMR STRUCTURE OF MOUSE DOPPEL 51-157 | | Descriptor: | PRION-LIKE PROTEIN | | Authors: | Mo, H, Moore, R.C, Cohen, F.E, Westaway, D, Prusiner, S.B, Wright, P.E, Dyson, H.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-03-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two different neurodegenerative diseases caused by proteins with similar structures.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1DO6
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE IN THE OXIDIZED STATE AT 2.0 ANGSTROM RESOLUTION | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Junior, F.E, Adams, M.W, Rees, D.C. | | Deposit date: | 1999-12-19 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
1DYZ
 
 | | OXIDISED AZURIN II FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | AZURIN II, COPPER (II) ION | | Authors: | Dodd, F.E, Abraham, Z.H.L, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2000-02-11 | | Release date: | 2000-07-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Oxidised and Reduced Azurin II from Alcaligenes Xylosoxidans at 1.75 Angstoms Resolution
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3G9Y
 
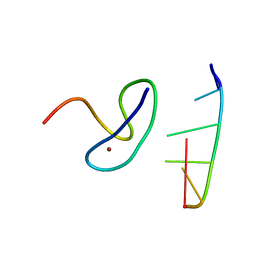 | | Crystal structure of the second zinc finger from ZRANB2/ZNF265 bound to 6 nt ssRNA sequence AGGUAA | | Descriptor: | RNA (5'-R(*AP*GP*GP*UP*AP*A)-3'), ZINC ION, Zinc finger Ran-binding domain-containing protein 2 | | Authors: | Loughlin, F.E, McGrath, A.P, Lee, M, Guss, J.M, Mackay, J.P. | | Deposit date: | 2009-02-15 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7AA4
 
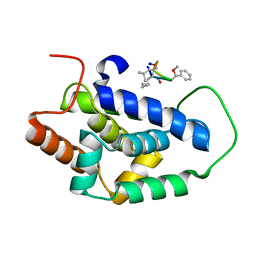 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7ABR
 
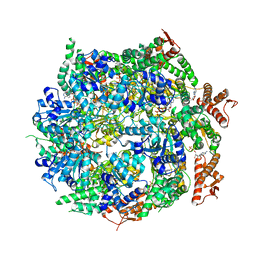 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
4C18
 
 | |
4ER4
 
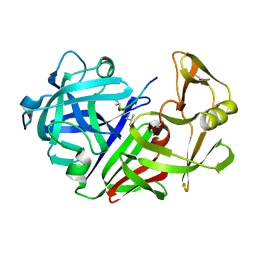 | | HIGH-RESOLUTION X-RAY ANALYSES OF RENIN INHIBITOR-ASPARTIC PROTEINASE COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, H-142 | | Authors: | Foundling, S.I, Watson, F.E, Szelke, M, Blundell, T.L. | | Deposit date: | 1991-01-05 | | Release date: | 1991-04-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High resolution X-ray analyses of renin inhibitor-aspartic proteinase complexes.
Nature, 327, 1987
|
|
4C8O
 
 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the aritificial base pair dNaM-d5SICS at the postinsertion site (sequence context 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*GP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*TP*TP*CP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*CP)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8K
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a partially closed complex with the artificial base pair d5SICS-dNaMTP | | Descriptor: | ((2R,3S,5R)-3-hydroxy-5-(3-methoxynaphthalen-2-yl)methyl-tetrahydrogen-triphosphate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*AP*C*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4C8L
 
 | | Binary complex of the large fragment of DNA polymerase I from Thermus Aquaticus with the artificial base pair dNaM-d5SICS at the postinsertion site (sequence context 1) | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*LHOP)-3', 5'-D(*AP*GP*BMNP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4FRN
 
 | | Crystal structure of the cobalamin riboswitch regulatory element | | Descriptor: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4FRG
 
 | | Crystal structure of the cobalamin riboswitch aptamer domain | | Descriptor: | Hydroxocobalamin, IRIDIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
5TY4
 
 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | Descriptor: | TGF-beta receptor type-2, mmTGF-b2-7m | | Authors: | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
