3HGY
 
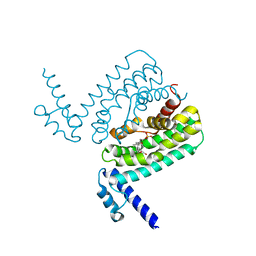 | | Crystal Structure of CmeR Bound to Taurocholic Acid | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: crystal structures of CmeR-bile acid complexes
To be Published
|
|
3K0I
 
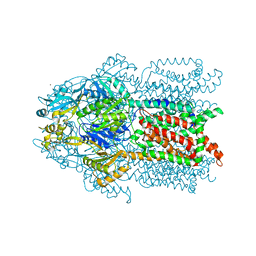 | | Crystal structure of Cu(I)CusA | | Descriptor: | COPPER (I) ION, Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.116 Å) | | Cite: | Crystal structure of CusA
To be Published
|
|
3K07
 
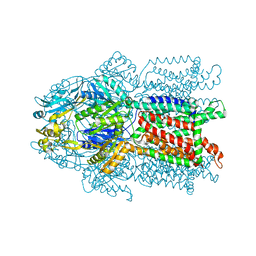 | | Crystal structure of CusA | | Descriptor: | Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
7KDT
 
 | |
7JZ6
 
 | |
7JZH
 
 | |
7K8D
 
 | |
7K8C
 
 | |
7K8A
 
 | |
7K8B
 
 | |
7K7M
 
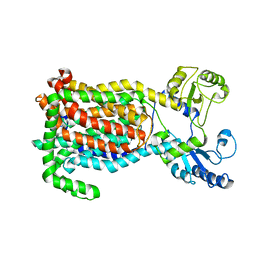 | | Crystal Structure of a membrane protein | | Descriptor: | Drug exporters of the RND superfamily-like protein, alpha-D-glucopyranose-(1-1)-6-O-decanoyl-alpha-D-glucopyranose | | Authors: | Su, C.-C. | | Deposit date: | 2020-09-23 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
3KSO
 
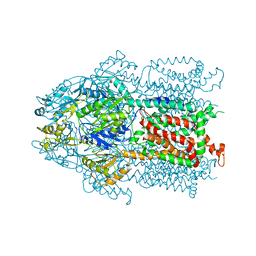 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | Cation efflux system protein cusA, SILVER ION | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.367 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
5C3C
 
 | |
3U0T
 
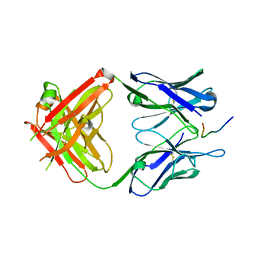 | | Fab-antibody complex | | Descriptor: | Amyloid beta A4 protein, ponezumab HC Fab, ponezumab LC Fab | | Authors: | LaPorte, S.L, Pons, J.P. | | Deposit date: | 2011-09-29 | | Release date: | 2012-01-11 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of C-terminal beta-Amyloid Peptide Binding by the Antibody Ponezumab for the Treatment of Alzheimer's Disease
J.Mol.Biol., 421, 2012
|
|
3KSS
 
 | | Structure and Mechanism of the Heavy Metal Transporter CusA | | Descriptor: | COPPER (I) ION, Cation efflux system protein cusA | | Authors: | Su, C.-C. | | Deposit date: | 2009-11-23 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport.
Nature, 467, 2010
|
|
3L7L
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (30 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Lovering, A.L, Strynadka, N.C.J. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L7J
 
 | |
3L7K
 
 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (15 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Strynadka, N.C.J, Lovering, A.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L7I
 
 | |
5D9R
 
 | | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6 | | Descriptor: | Protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic | | Authors: | Radhakrishnan, A, Kumar, N, Su, C.-C, Chou, T.-H, Yu, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6.
Protein Sci., 25, 2016
|
|
3L7M
 
 | |
5D1R
 
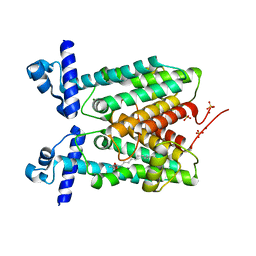 | | Crystal structure of Mycobacterium tuberculosis Rv1816 transcriptional regulator. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Rv1816 transcriptional regulator, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
3SH2
 
 | |
3SGY
 
 | |
5D18
 
 | | Crystal structure of Mycobacterium tuberculosis Rv0302, form I | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis transcriptional regulator Rv0302.
Protein Sci., 2015
|
|
