1J53
 
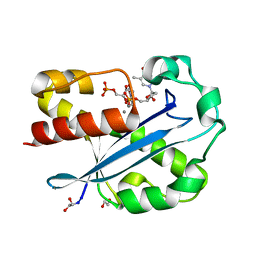 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1JAW
 
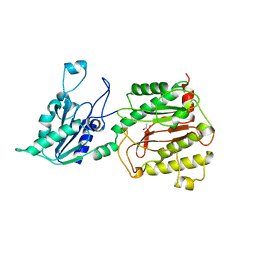 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1J54
 
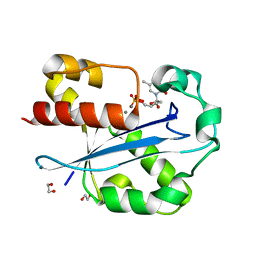 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1JWE
 
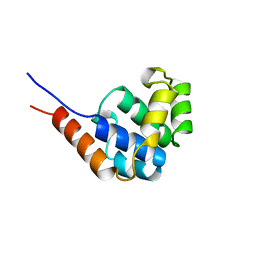 | | NMR Structure of the N-Terminal Domain of E. Coli Dnab Helicase | | Descriptor: | PROTEIN (DNAB HELICASE) | | Authors: | Weigelt, J, Brown, S.E, Miles, C.S, Dixon, N.E, Otting, G. | | Deposit date: | 1999-01-22 | | Release date: | 1999-01-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer.
Structure Fold.Des., 7, 1999
|
|
2PII
 
 | | PII, GLNB PRODUCT | | Descriptor: | PII | | Authors: | Carr, P.D, Cheah, E, Suffolk, P.M, Ollis, D.L. | | Deposit date: | 1995-05-02 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the signal transduction protein from Escherichia coli at 1.9 A.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PIL
 
 | |
3X1O
 
 | | Crystal structure of the ROQ domain of human Roquin | | Descriptor: | IODIDE ION, Roquin-1 | | Authors: | Ose, T, Verma, A, Cockburn, J.B, Berrow, N.S, Alderton, D, Stuart, D, Owens, R.J, Jones, E.Y. | | Deposit date: | 2014-11-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis
Nat Commun, 6, 2015
|
|
6CIX
 
 | |
6CEJ
 
 | |
4XR0
 
 | |
4XR1
 
 | |
4XR3
 
 | |
4XR2
 
 | |
7SSG
 
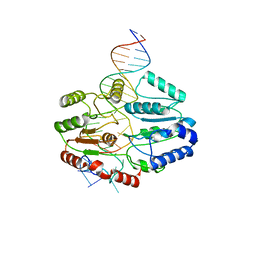 | | Mfd DNA complex | | Descriptor: | DNA (5'-D(P*TP*GP*GP*CP*GP*GP*CP*GP*AP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*GP*CP*CP*TP*CP*GP*CP*TP*GP*CP*CP*A)-3'), Transcription-repair-coupling factor | | Authors: | Oakley, A.J, Xu, Z.-Q. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Mechanism of transcription modulation by the transcription-repair coupling factor.
Nucleic Acids Res., 50, 2022
|
|
2NUL
 
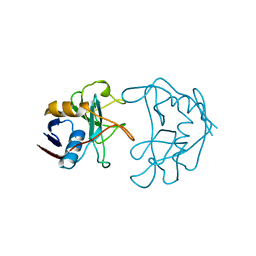 | | PEPTIDYLPROLYL ISOMERASE FROM E. COLI | | Descriptor: | PEPTIDYLPROLYL ISOMERASE | | Authors: | Edwards, K.J, Ollis, D.L. | | Deposit date: | 1996-11-14 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cytoplasmic Escherichia coli peptidyl-prolyl isomerase: evidence for decreased mobility of loops upon complexation.
J.Mol.Biol., 271, 1997
|
|
6AP4
 
 | |
6AMQ
 
 | |
6CIV
 
 | |
6CBI
 
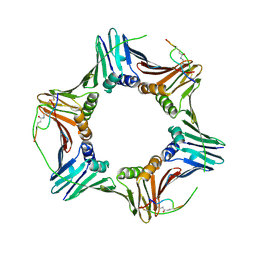 | | PCNA in complex with inhibitor | | Descriptor: | GLY-ARG-LYS-ARG-ARG-GLN-DAB-SER-MET-THR-GLU-PHE-TYR-HIS, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Bruning, J.B, Wegener, K.L. | | Deposit date: | 2018-02-03 | | Release date: | 2018-07-04 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Chemistry, 24, 2018
|
|
6AMS
 
 | |
4MJR
 
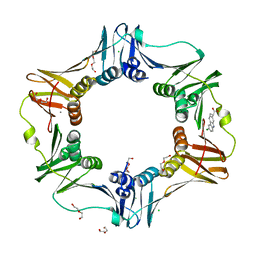 | | E. coli sliding clamp in complex with (S)-Carprofen | | Descriptor: | (2S)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
4MJP
 
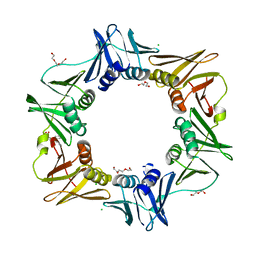 | | E. coli sliding clamp in complex with (R)-Vedaprofen | | Descriptor: | (2R)-2-(4-cyclohexylnaphthalen-1-yl)propanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
4MJQ
 
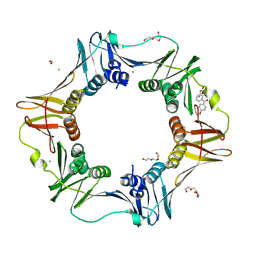 | | E. coli sliding clamp in complex with Bromfenac | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs.
Chem.Biol., 21, 2014
|
|
4N95
 
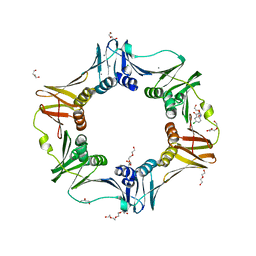 | | E. coli sliding clamp in complex with 5-chloroindoline-2,3-dione | | Descriptor: | 5-chloro-1H-indole-2,3-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N94
 
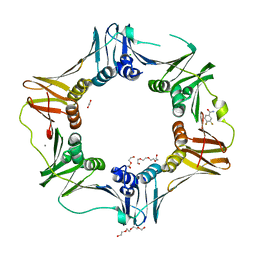 | | E. coli sliding clamp in complex with 3,4-difluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 3,4-difluorobenzamide, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
