5V56
 
 | | 2.9A XFEL structure of the multi-domain human smoothened receptor (with E194M mutation) in complex with TC114 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN MONONUCLEOTIDE, N-methyl-N-[1-[4-(2-methylpyrazol-3-yl)phthalazin-1-yl]piperidin-4-yl]-4-nitro-2-(trifluoromethyl)benzamide, ... | | Authors: | Zhang, X, Zhao, F, Wu, Y, Yang, J, Han, G.W, Zhao, S, Ishchenko, A, Ye, L, Lin, X, Ding, K, Dharmarajan, V, Griffin, P.R, Gati, C, Nelson, G, Hunter, M.S, Hanson, M.A, Cherezov, V, Stevens, R.C, Tan, W, Tao, H, Xu, F. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand.
Nat Commun, 8, 2017
|
|
3JB6
 
 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase, VP1 CSP, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-02 | | Release date: | 2015-10-28 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
3JB7
 
 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (4.001 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
5FDP
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
5FDX
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution. | | Descriptor: | 1,2-ETHANEDIOL, 3-[(4-methylpiperazin-1-yl)methyl]-~{N}-[(4~{R})-4-methyl-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinolin-7-yl]-5-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Chalk, R, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution.
To Be Published
|
|
2KBN
 
 | | Solution NMR structure of the OB domain (residues 67-166) of MM0293 from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR214a. | | Descriptor: | Conserved protein | | Authors: | Ramelot, T.A, Ding, K, Magliqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the OB domain of mm0293 from the archea methanosarcina mazei
To be Published
|
|
2KCD
 
 | | Solution NMR structure of SSP0047 from Staphylococcus saprophyticus. Northeast Structural Genomics Consortium Target SyR6. | | Descriptor: | Uncharacterized protein SSP0047 | | Authors: | Ramelot, T.A, Ding, K, Chen, C.X, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-10 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SSP0047 from Staphylococcus saprophyticus. Northeast Structural Genomics Consortium Target SyR6.
To be Published
|
|
2KEN
 
 | | Solution NMR structure of the OB domain (residues 67-166) of MM0293 from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR214a. | | Descriptor: | Conserved protein | | Authors: | Ramelot, T.A, Ding, K, Magliqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-10 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the OB domain of MM0293 from the archea Methanosarcina mazei
To be Published
|
|
2K75
 
 | | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum. Northeast Structural Genomics Consortium target TaR80b. | | Descriptor: | uncharacterized protein Ta0387 | | Authors: | Ramelot, T.A, Ding, K, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-01 | | Release date: | 2008-08-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum.
To be Published
|
|
4PW2
 
 | | Crystal structure of D-glucuronyl C5 epimerase | | Descriptor: | CITRIC ACID, D-glucuronyl C5 epimerase B | | Authors: | Ke, J, Qin, Y, Gu, X, Brunzelle, J.S, Xu, H.E, Ding, K. | | Deposit date: | 2014-03-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
6BQH
 
 | | Crystal structure of 5-HT2C in complex with ritanserin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, 6-(2-{4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl}ethyl)-7-methyl-5H-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ... | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
3TZR
 
 | | Structure of a Riboswitch-like RNA-ligand complex from the Hepatitis C Virus Internal Ribosome Entry Site | | Descriptor: | (8R)-8-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-1,7,8,9-tetrahydrochromeno[5,6-d]imidazol-2-amine, 5'-R(*CP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*CP*C)-3', 5'-R(*GP*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*UP*CP*GP*G)-3', ... | | Authors: | Dibrov, S.M, Ding, K, Brunn, N, Parker, M.A, Bergdahl, B.M, Wyles, D.L, Hermann, T. | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | Structure of a Riboswitch in the Hepatitis C Virus Internal Ribosome Entry Site
To be Published
|
|
5XRA
 
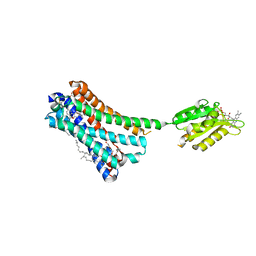 | | Crystal structure of the human CB1 in complex with agonist AM11542 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6aR,10aR)-3-(8-bromanyl-2-methyl-octan-2-yl)-6,6,9-trimethyl-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, CHOLESTEROL, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1
Nature, 547, 2017
|
|
5XR8
 
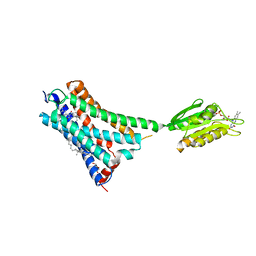 | | Crystal structure of the human CB1 in complex with agonist AM841 | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, CHOLESTEROL, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1.
Nature, 547, 2017
|
|
2HJQ
 
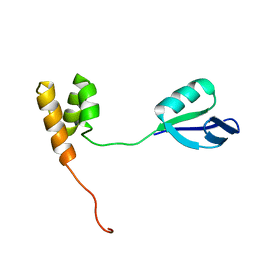 | | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449 | | Descriptor: | Hypothetical protein yqbF | | Authors: | Ding, K, Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-30 | | Release date: | 2007-06-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus Subtilis Protein YqbF, Northeast Structural Genomics Target SR449
TO BE PUBLISHED
|
|
7ER2
 
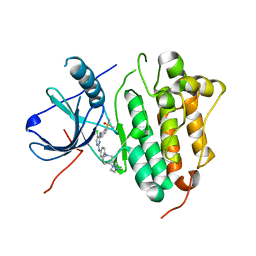 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with LS_2_40 | | Descriptor: | 5-chloranyl-N2-[3-chloranyl-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-N4-(2-dimethylphosphorylphenyl)pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2021-05-05 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Optimization of Brigatinib as New Wild-Type Sparing Inhibitors of EGFR T790M/C797S Mutants.
Acs Med.Chem.Lett., 13, 2022
|
|
6GWR
 
 | | Structure of the kinase domain of human DDR1 in complex with a potent and selective inhibitor of DDR1 and DDR2 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-benzamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 3-(Imidazo[1,2- a]pyrazin-3-ylethynyl)-4-isopropyl- N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a Dual Inhibitor of Discoidin Domain Receptors 1 and 2.
J. Med. Chem., 61, 2018
|
|
7VRE
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 | | Descriptor: | 5-chloranyl-N-[5-chloranyl-2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-4-(1-ethylsulfonylindol-3-yl)pyrimidin-2-amine, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
7VRA
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HC5476 | | Descriptor: | 25-chloro-11-(ethylsulfonyl)-44-morpholino-11H-5,12-dioxa-3-aza-1(3,6)-indola-2(4,2)-pyrimidina-4(1,3)-benzenacyclododecaphane, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
6NVH
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVJ
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVL
 
 | | FGFR1 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 1, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVG
 
 | | FGFR4 complex with N-(3,5-dichloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)phenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3,5-dichloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)phenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVK
 
 | | FGFR4 complex with BLU-554, N-((3S,4S)-3-((6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl)amino)tetrahydro-2H-pyran-4-yl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3S,4S)-3-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl]amino}oxan-4-yl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
