8OHT
 
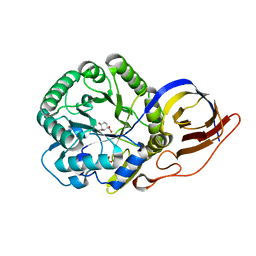 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHQ
 
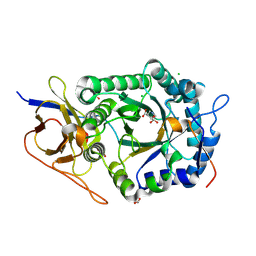 | | Crystal structure of human heparanase in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Davies, G.J, Overkleeft, H.S, Armstrong, Z, Yurong, C. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHX
 
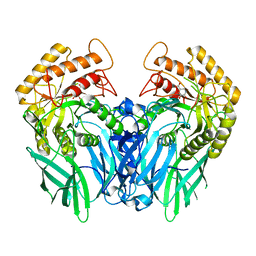 | | Crystal structure of Beta-glucuronidase from Escherichia coli in complex with siastatin B derived inhibitor | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OGX
 
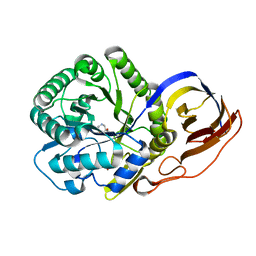 | | Beta-glucuronidase from Acidobacterium capsulatum in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, PHOSPHATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHV
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured 3-geminal diol iminosugar inhibitor | | Descriptor: | (3~{S},4~{R})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHW
 
 | | Crystal structure of heparanase from Burkholderia pseudomallei in complex with siastatin B derived inhibitor | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, Glycoside hydrolase family 44 domain-containing protein | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHU
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured isofagamine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8PEJ
 
 | | CjGH35 with a Galactosidase Activity-Based Probe | | Descriptor: | (1~{S},2~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-06-14 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The development of a broad-spectrum retaining beta-exo-galactosidase activity-based probe.
Org.Biomol.Chem., 21, 2023
|
|
8Q59
 
 | | Crystal structure of metal-dependent class II sulfofructose phosphate aldolase from Yersinia aldovae in complex with sulfofructose phosphate (YaSqiA-Zn-SFP) | | Descriptor: | (3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-oxidanylidene-6-phosphonooxy-hexane-1-sulfonic acid, SODIUM ION, Tagatose-1,6-bisphosphate aldolase kbaY, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8Q5A
 
 | | Crystal structure of metal-dependent class II sulfofructosephosphate aldolase from Hafnia paralvei HpSqiA-Zn in complex with dihydroxyacetone phosphate (DHAP) | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Ketose-bisphosphate aldolase, ZINC ION | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8Q58
 
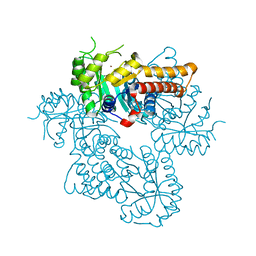 | |
8Q57
 
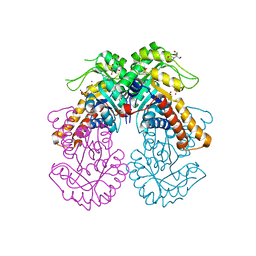 | | Crystal structure of class II SFP aldolase from Yersinia aldovae (YaSqiA-Zn-SO4) with bound sulfate ions | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SODIUM ION, SULFATE ION, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
7DIF
 
 | | GH127 beta-L-arabinofuranosidase HypBA1 covalently complexed with beta-L-arabinofuranose-configured cyclophellitol at 1.75-angstrom resolution | | Descriptor: | (1S,2S,3R,4R)-3-(hydroxymethyl)cyclopentane-1,2,4-triol, Non-reducing end beta-L-arabinofuranosidase, POTASSIUM ION, ... | | Authors: | Amaki, S, McGregor, N.G.S, Arakawa, T, Yamada, C, Borlandelli, V, Overkleeft, H.S, Davies, G.J, Fushinobu, S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8P0L
 
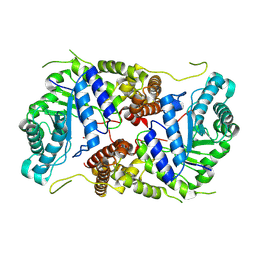 | | Crystal structure of human O-GlcNAcase in complex with an S-linked CKII peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Protein O-GlcNAcase, ... | | Authors: | Males, A, Davies, G.J, Calvelo, M, Alteen, M.G, Vocadlo, D.J, Rovira, C. | | Deposit date: | 2023-05-10 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human O -GlcNAcase Uses a Preactivated Boat-skew Substrate Conformation for Catalysis. Evidence from X-ray Crystallography and QM/MM Metadynamics.
Acs Catalysis, 13, 2023
|
|
8OZ1
 
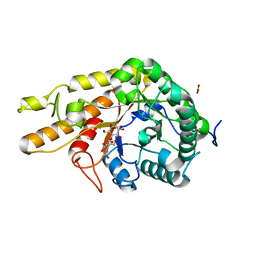 | | CjCel5D endo-xyloglucanase bounc to CB665 covalent inhibitor | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, BORIC ACID, Cellulase, ... | | Authors: | McGregor, N.G.S, de Boer, C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8QC5
 
 | |
8QC8
 
 | |
8QC6
 
 | |
8QC2
 
 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
8QC3
 
 | |
8R06
 
 | | CRYSTAL STRUCTURE OF THERMOANAEROBACTERIUM XYLANOLYTICUM GH116 BETA-GLUCOSIDASE WITH A COVALENTLY BOUND CYCLOPHELLITOL AZIRIDINE | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},5~{R},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Davies, G.J, Breen, I.Z. | | Deposit date: | 2023-10-30 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOANAEROBACTERIUM XYLOLYTICUM GH116 BETA-GLUCOSIDASE WITH A COVALENTLY BOUND CYCLOPHELLITOL AZIRIDINE
JACS, 40, 2017
|
|
8QF8
 
 | | GH146 beta-L-arabinofuranosidase from Bacteroides thetaioatomicron in complex with beta-l-arabinofurano cyclophellitol aziridine | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, (1~{S},2~{S},3~{S},4~{S},5~{S})-4-(hydroxymethyl)-6-azabicyclo[3.1.0]hexane-2,3-diol, Glycosyl hydrolase, ... | | Authors: | Borlandelli, V, Offen, W, Moroz, O.V, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-04 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
8QF2
 
 | | Beta-L-Arabinofurano-cyclitol Aziridines are Cysteine-directed Broad-spectrum Inhibitors and Activity-based Probes for Retaining Beta-L-arabinofuranosidases | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Borlandelli, V, Offen, W.A, Moroz, O, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
7YZU
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with SQMe | | Descriptor: | Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-methoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Davies, G.J. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7YZS
 
 | |
