1R8U
 
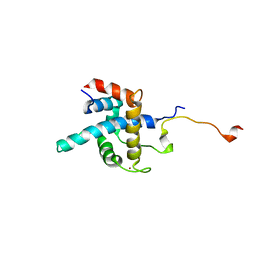 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
2AGH
 
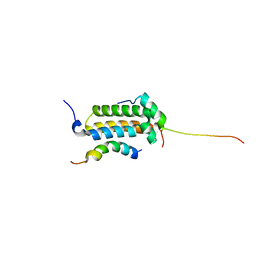 | | Structural basis for cooperative transcription factor binding to the CBP coactivator | | Descriptor: | Crebbp protein, Myb proto-oncogene protein, Zinc finger protein HRX | | Authors: | De Guzman, R.N, Goto, N.K, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cooperative Transcription Factor Binding to the CBP Coactivator
J.Mol.Biol., 355, 2006
|
|
1A1T
 
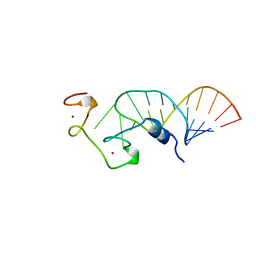 | | STRUCTURE OF THE HIV-1 NUCLEOCAPSID PROTEIN BOUND TO THE SL3 PSI-RNA RECOGNITION ELEMENT, NMR, 25 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, SL3 STEM-LOOP RNA, ZINC ION | | Authors: | De Guzman, R.N, Wu, Z.R, Stalling, C.C, Pappalardo, L, Borer, P.N, Summers, M.F. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element.
Science, 279, 1998
|
|
1U2N
 
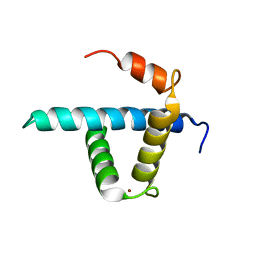 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
1F81
 
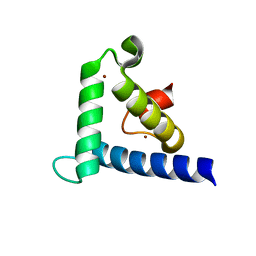 | | SOLUTION STRUCTURE OF THE TAZ2 DOMAIN OF THE TRANSCRIPTIONAL ADAPTOR PROTEIN CBP | | Descriptor: | CREB-BINDING PROTEIN, ZINC ION | | Authors: | De Guzman, R.N, Liu, H.L, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-28 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP.
J.Mol.Biol., 303, 2000
|
|
1L8C
 
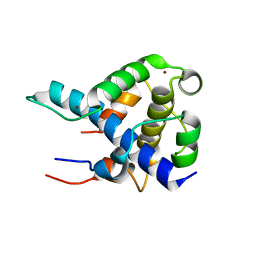 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1SB0
 
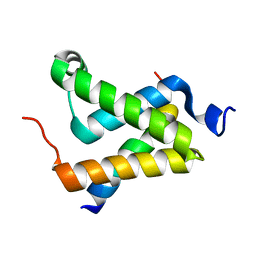 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
1MFS
 
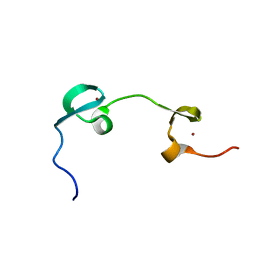 | | DYNAMICAL BEHAVIOR OF THE HIV-1 NUCLEOCAPSID PROTEIN; NMR, 30 STRUCTURES | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Turner, B.G, De Guzman, R.N, Lee, B.M, Tjandra, N. | | Deposit date: | 1998-04-01 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dynamical behavior of the HIV-1 nucleocapsid protein.
J.Mol.Biol., 279, 1998
|
|
1XJH
 
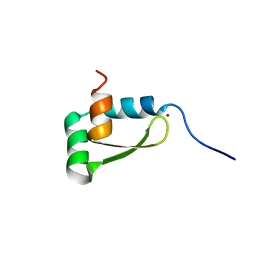 | | NMR structure of the redox switch domain of the E. coli Hsp33 | | Descriptor: | 33 kDa chaperonin, ZINC ION | | Authors: | Won, H.S, Low, L.Y, De Guzman, R.N, Martinez-Yamout, M.A, Jakob, U, Dyson, H.J. | | Deposit date: | 2004-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Zinc-dependent Redox Switch Domain of the Chaperone Hsp33 has a Novel Fold
J.Mol.Biol., 341, 2004
|
|
2G0U
 
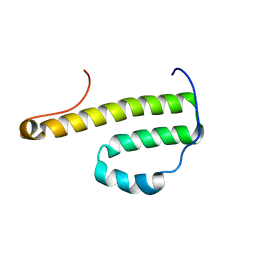 | | Solution Structure of Monomeric BsaL, the Type III Secretion Needle Protein of Burkholderia pseudomallei | | Descriptor: | type III secretion system needle protein | | Authors: | Zhang, L, Wang, Y, Picking, W.L, Picking, W.D, De Guzman, R.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Monomeric BsaL, the Type III Secretion Needle Protein of Burkholderia pseudomallei.
J.Mol.Biol., 359, 2006
|
|
2K48
 
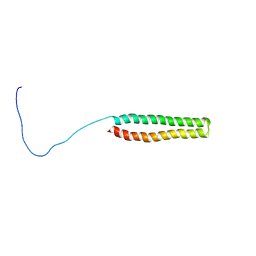 | | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Wang, Y, Boudreaux, D.M, Estrada, D.F, Egan, C.W, St Jeor, S.C, De Guzman, R.N. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein.
J.Biol.Chem., 283, 2008
|
|
2JOW
 
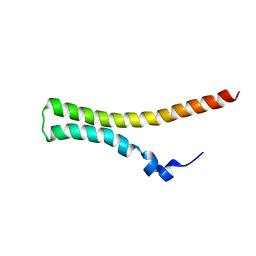 | | Differences in the electrostatic surfaces of the type III secretion needle proteins | | Descriptor: | Protein prgI | | Authors: | Wang, Y, Ouellette, A.N, Egan, C.W, De Guzman, R.N. | | Deposit date: | 2007-04-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differences in the Electrostatic Surfaces of the Type III Secretion Needle Proteins PrgI, BsaL, and MxiH
J.Mol.Biol., 371, 2007
|
|
2K9H
 
 | | The hantavirus glycoprotein G1 tail contains a dual CCHC-type classical zinc fingers | | Descriptor: | Glycoprotein, ZINC ION | | Authors: | Estrada, D.F, Boudreaux, D.M, Zhong, D, St Jeor, S.C, De Guzman, R.N. | | Deposit date: | 2008-10-13 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Hantavirus Glycoprotein G1 Tail Contains Dual CCHC-type Classical Zinc Fingers.
J.Biol.Chem., 284, 2009
|
|
2L7X
 
 | |
1ESY
 
 | |
1F6U
 
 | | NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop sl2 of the psi-RNA packaging signal. Implications for genome recognition | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, HIV-1 STEM-LOOP SL2 FROM PSI-RNA PACKAGING, ZINC ION | | Authors: | Amarasinghe, G.K, De Guzman, R.N, Turner, R.B, Chancellor, K.J, Summers, M.F. | | Deposit date: | 2000-06-23 | | Release date: | 2000-10-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition.
J.Mol.Biol., 301, 2000
|
|
1DSV
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (C-TERMINAL ZINC FINGER) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1DSQ
 
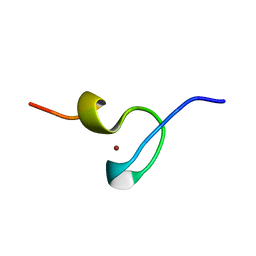 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (ZINC FINGER 1) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
4JBU
 
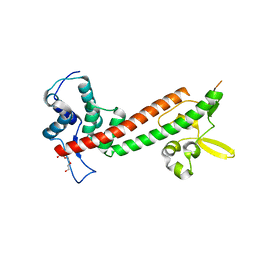 | | 1.65A structure of the T3SS tip protein LcrV (G28-D322, C273S) from Yersinia pestis | | Descriptor: | TETRAETHYLENE GLYCOL, Virulence-associated V antigen | | Authors: | Lovell, S, Chaudhury, S, Battaile, K.P, Plano, G, De Guzman, R.N. | | Deposit date: | 2013-02-20 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Yersinia pestis tip protein LcrV refined to 1.65A resolution
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6CJD
 
 | |
3O01
 
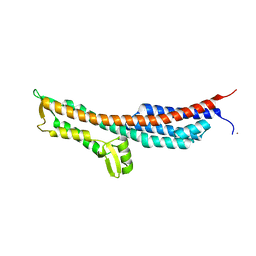 | | The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
3NZZ
 
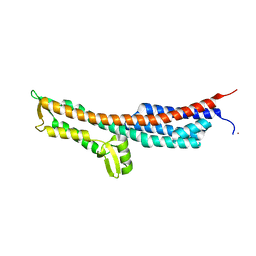 | | Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD | | Descriptor: | Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
3O00
 
 | | Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD-C244S | | Descriptor: | Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
3O02
 
 | | The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Chenodeoxycholate | | Descriptor: | CHENODEOXYCHOLIC ACID, Cell invasion protein sipD, NICKEL (II) ION | | Authors: | Chatterjee, S, Zhong, D, Nordhues, B.A, Battaile, K.P, Lovell, S, DeGuzman, R.N. | | Deposit date: | 2010-07-18 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate.
Protein Sci., 20, 2011
|
|
