2Z5D
 
 | | Human ubiquitin-conjugating enzyme E2 H | | Descriptor: | SODIUM ION, Ubiquitin-conjugating enzyme E2 H | | Authors: | Bochkarev, A, Cui, H, Walker, J.R, Newman, E.M, Mackenzie, F, Battaile, K.P, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
MOL.CELL PROTEOMICS, 11, 2012
|
|
3HJZ
 
 | | The structure of an aldolase from Prochlorococcus marinus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3I9S
 
 | | Structure from the mobile metagenome of V.cholerae. Integron cassette protein VCH_CASS6 | | Descriptor: | CHLORIDE ION, Integron cassette protein, SULFATE ION | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Chang, C, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-13 | | Release date: | 2009-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure from the mobile metagenome of V.cholerae. Integron cassette protein VCH_CASS6
To be Published
|
|
3JRT
 
 | | Structure from the mobile metagenome of V. paracholerae: Integron cassette protein Vpc_cass2 | | Descriptor: | Integron cassette protein Vpc_cass2 | | Authors: | Harrop, S.J, Deshpande, C, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Cuff, M, Edwards, A, Savchenko, A, Joachimiak, A, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
5VL3
 
 | | CD22 d1-d3 in complex with therapeutic Fab Epratuzumab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell receptor CD22, Epratuzumab Fab Heavy Chain, ... | | Authors: | Sicard, T, Ereno-Orbea, J, Julien, J.P. | | Deposit date: | 2017-04-24 | | Release date: | 2017-10-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
5VKJ
 
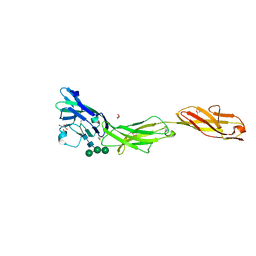 | | Crystal structure of human CD22 Ig domains 1-3 | | Descriptor: | B-cell receptor CD22, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Julien, J.P, Ereno-Orbea, J, Sicard, T. | | Deposit date: | 2017-04-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
4Q3K
 
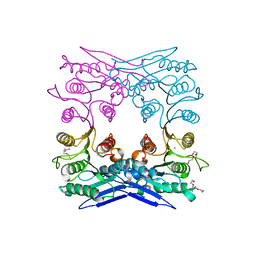 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3N
 
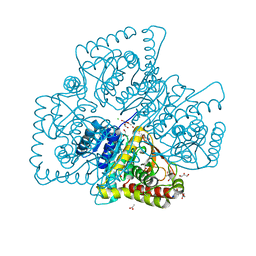 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3O
 
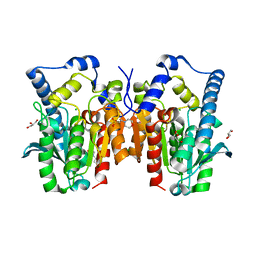 | | Crystal structure of MGS-MT1, an alpha/beta hydrolase enzyme from a Lake Matapan deep-sea metagenome library | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4Q3M
 
 | | Crystal structure of MGS-M4, an aldo-keto reductase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | MGS-M4, SODIUM ION, SULFATE ION | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
3FY6
 
 | | Structure from the mobile metagenome of V. Cholerae. Integron cassette protein VCH_CASS3 | | Descriptor: | Integron cassette protein | | Authors: | Deshpande, C.N, Sureshan, V, Harrop, S.J, Boucher, Y, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Tan, K, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-21 | | Release date: | 2009-02-10 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
6MR3
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans | | Descriptor: | CHLORIDE ION, Putative competence-damage inducible protein | | Authors: | Stogios, P.J, Cuff, M, Xu, X, Cui, H, Di Leo, R, Yim, V, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-10-11 | | Release date: | 2018-10-24 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the competence-damaged protein (CinA) superfamily protein from Streptococcus mutans
To Be Published
|
|
7EKK
 
 | | Anti-HIV-1 broadly neutralizing antibody delta-loop 4E10 modified with pyrene acetamide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
7EKB
 
 | | Crystal structure of 4E10 modified with pyrene acetamide | | Descriptor: | ACETATE ION, Fab region of the heavy chain of broadly neutralizing antibody anti-HIV-1 4E10, Fab region of the light chain of the broadly neutralizing anti-HIV-1 antibody 4E10, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
