2OFP
 
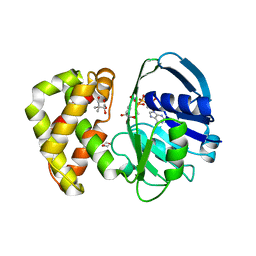 | | Crystal structure of Escherichia coli ketopantoate reductase in a ternary complex with NADP+ and pantoate | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETATE ION, Ketopantoate reductase, ... | | Authors: | Ciulli, A, Chirgadze, D.Y, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Ketopantoate Reductase in a Ternary Complex with NADP+ and Pantoate Bound: SUBSTRATE RECOGNITION, CONFORMATIONAL CHANGE, AND COOPERATIVITY.
J.Biol.Chem., 282, 2007
|
|
2QM4
 
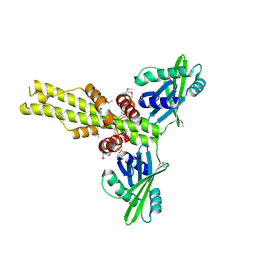 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
6ZFP
 
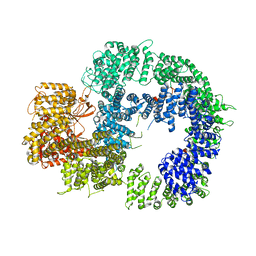 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
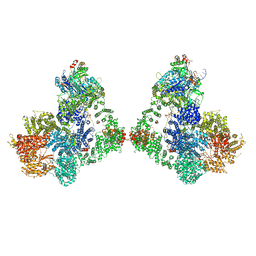 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
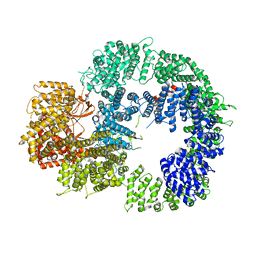 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
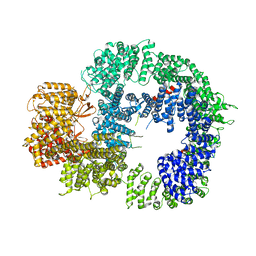 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
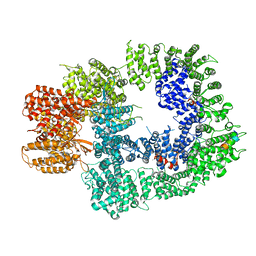 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHA
 
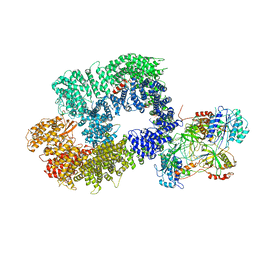 | | Cryo-EM structure of DNA-PK monomer | | Descriptor: | DNA, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH8
 
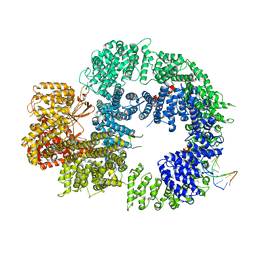 | | Cryo-EM structure of DNA-PKcs:DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*T)-3'), DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1URF
 
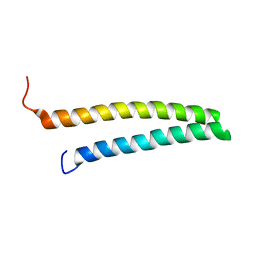 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
2FN0
 
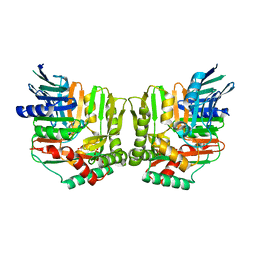 | | Crystal structure of Yersinia enterocolitica salicylate synthase (Irp9) | | Descriptor: | ACETATE ION, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kerbarh, O, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2006-01-10 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Yersinia enterocolitica Salicylate Synthase and its Complex with the Reaction Products Salicylate and Pyruvate.
J.Mol.Biol., 357, 2006
|
|
2FN1
 
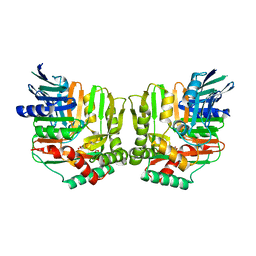 | | Crystal structures of Yersinia enterocolitica salicylate synthase (Irp9) in complex with the reaction products salicylate and pyruvate | | Descriptor: | 2-HYDROXYBENZOIC ACID, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kerbarh, O, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2006-01-10 | | Release date: | 2006-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Yersinia enterocolitica Salicylate Synthase and its Complex with the Reaction Products Salicylate and Pyruvate.
J.Mol.Biol., 357, 2006
|
|
2HE0
 
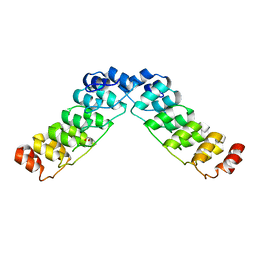 | | Crystal structure of a human Notch1 ankyrin domain mutant | | Descriptor: | 1,2-ETHANEDIOL, Notch1 preproprotein variant | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Martinez Arias, A, Blundell, T.L. | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human Notch1 ankyrin domain mutant
TO BE PUBLISHED
|
|
2J66
 
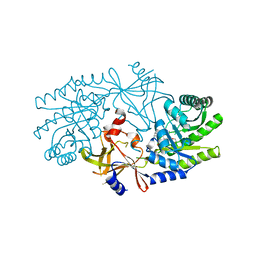 | | Structural characterisation of BtrK decarboxylase from butirosin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, BTRK, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Popovic, B, Li, Y, Chirgadze, D.Y, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterisation of Btrk Decarboxylase from Bacillus Circulans Butirosin Biosynthesis
To be Published
|
|
1GMN
 
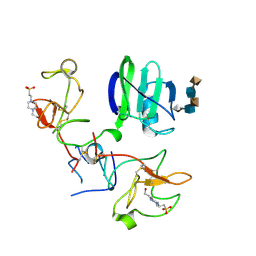 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1GMO
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
1YYH
 
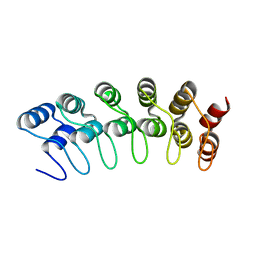 | | Crystal structure of the human Notch 1 ankyrin domain | | Descriptor: | Notch 1, ankyrin domain | | Authors: | Ehebauer, M.T, Chirgadze, D.Y, Hayward, P, Martinez-Arias, A, Blundell, T.L. | | Deposit date: | 2005-02-25 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | High-resolution crystal structure of the human Notch 1 ankyrin domain
Biochem.J., 392, 2005
|
|
1Z56
 
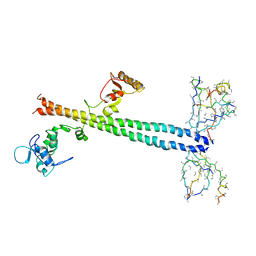 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
2ASU
 
 | |
2C81
 
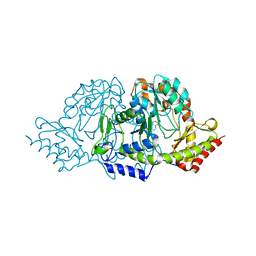 | | Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis. | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLUTAMINE-2-DEOXY-SCYLLO-INOSOSE AMINOTRANSFERASE | | Authors: | Popovic, B, Tang, X, Chirgadze, D.Y, Huang, F, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2005-11-30 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis.
Proteins, 65, 2006
|
|
2C7T
 
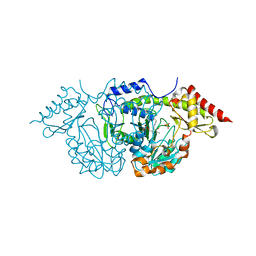 | | CRYSTAL STRUCTURE OF THE PLP-BOUND FORM OF BTRR, A DUAL FUNCTIONAL AMINOTRANSFERASE INVOLVED IN BUTIROSIN BIOSYNTHESIS. | | Descriptor: | GLUTAMINE-2-DEOXY-SCYLLO-INOSOSE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Popovic, B, Tang, X, Chirgadze, D.Y, Huang, F, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2005-11-29 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis.
Proteins, 65, 2006
|
|
7AUA
 
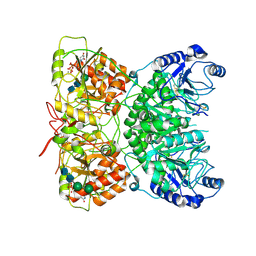 | | Cryo-EM structure of human exostosin-like 3 (EXTL3) in complex with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, MANGANESE (II) ION, ... | | Authors: | Wilson, L.F.L, Dendooven, T, Hardwick, S.W, Chirgadze, D.Y, Luisi, B.F, Logan, D.T, Mani, K, Dupree, P. | | Deposit date: | 2020-11-02 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.
Nat Commun, 13, 2022
|
|
7AU2
 
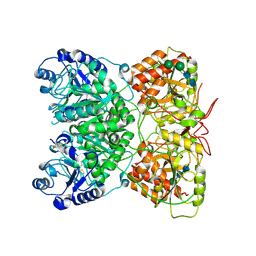 | | Cryo-EM structure of human exostosin-like 3 (EXTL3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wilson, L.F.L, Dendooven, T, Hardwick, S.W, Chirgadze, D.Y, Luisi, B.F, Logan, D.T, Mani, K, Dupree, P. | | Deposit date: | 2020-11-02 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.
Nat Commun, 13, 2022
|
|
7B1B
 
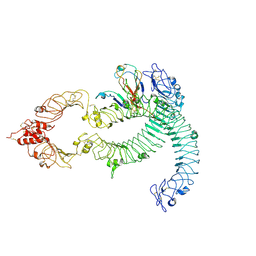 | | Cryo-EM of Aedes Aegypti Toll5A dimer bound to Spz1C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL013433-PA, ... | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
7B1C
 
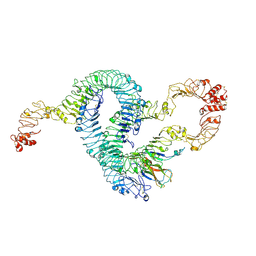 | | Cryo-EM of Aedes Aegypti Toll5A trimer bound to Spz1C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AAEL013433-PA, ... | | Authors: | Gangloff, M, Hardwick, S.W, Chirgadze, D.Y. | | Deposit date: | 2020-11-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun, 13, 2022
|
|
