5IWF
 
 | |
5IVF
 
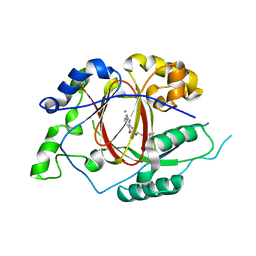 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | 分子名称: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.683 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVV
 
 | |
5IVJ
 
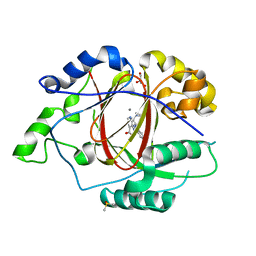 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | 分子名称: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-21 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.569 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVY
 
 | |
5IVE
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N8 ( 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile) | | 分子名称: | 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, MANGANESE (II) ION | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.783 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IW0
 
 | |
2NOG
 
 | |
5SZX
 
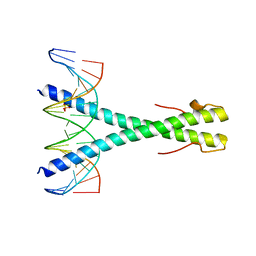 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5T0U
 
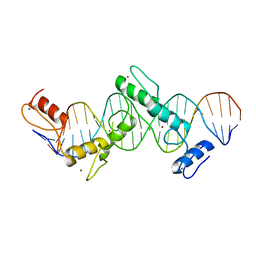 | | CTCF ZnF2-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5T00
 
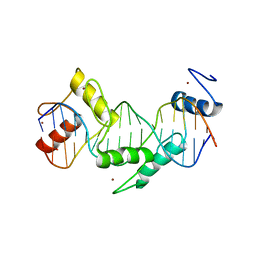 | | Human CTCF ZnF3-7 and methylated DNA complex | | 分子名称: | DNA (5'-GCCAGCAGGGGG(5CM)GCTA-3'), DNA (5'-TAG(5CM)GCCCCCTGCTGGC-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5T01
 
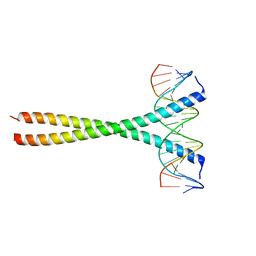 | | Human c-Jun DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*GP*AP*(5CM)P*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*GP*TP*CP*CP*AP*T)-3'), Transcription factor AP-1 | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5V3G
 
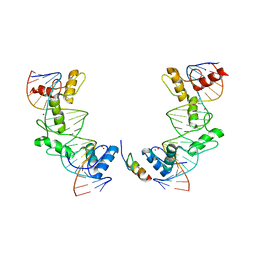 | | PRDM9-allele-C ZnF8-13 | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*CP*AP*AP*CP*GP*CP*TP*CP*AP*CP*TP*GP*GP*GP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*CP*CP*AP*GP*TP*GP*AP*GP*CP*GP*TP*TP*GP*CP*CP*C)-3'), PR domain zinc finger protein 9, ... | | 著者 | Patel, A, Cheng, X. | | 登録日 | 2017-03-07 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.416 Å) | | 主引用文献 | Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence.
J. Biol. Chem., 292, 2017
|
|
7C7R
 
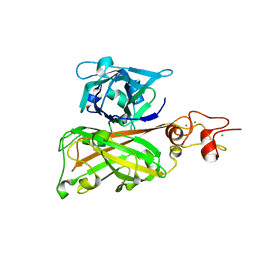 | | Biofilm associated protein - B domain | | 分子名称: | Biofilm-associated surface protein, CALCIUM ION | | 著者 | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | 登録日 | 2020-05-26 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-07-28 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
7C7U
 
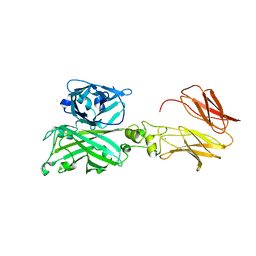 | | Biofilm associated protein - BSP domain | | 分子名称: | Biofilm-associated surface protein, CALCIUM ION | | 著者 | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | 登録日 | 2020-05-26 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
5V3M
 
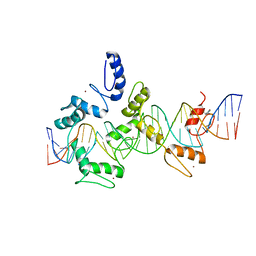 | |
5V3J
 
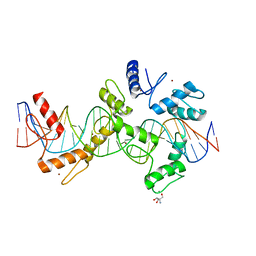 | | mouseZFP568-ZnF1-10 in complex with DNA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (26-MER), MAGNESIUM ION, ... | | 著者 | Patel, A, Cheng, X. | | 登録日 | 2017-03-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.064 Å) | | 主引用文献 | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
5UND
 
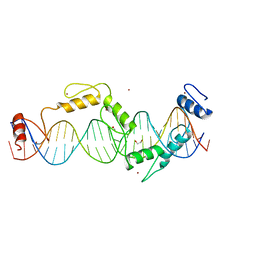 | |
5WJQ
 
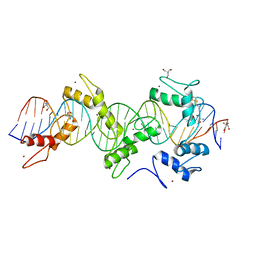 | | mouseZFP568-ZnF2-11 in complex with DNA | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (28-MER), ZINC ION, ... | | 著者 | Patel, A, Cheng, X. | | 登録日 | 2017-07-24 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.794 Å) | | 主引用文献 | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
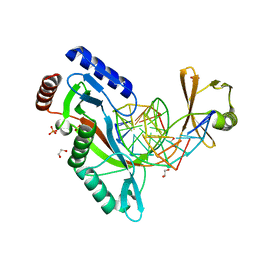
6UKG
 
 | |
6UKF
 
 | |
6UKI
 
 | |
6UKE
 
 | |
6UKH
 
 | |
6V62
 
 | | SETD3 double mutant (N255F/W273A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | 分子名称: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | 著者 | Dai, S, Horton, J.R, Cheng, X. | | 登録日 | 2019-12-04 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | An engineered variant of SETD3 methyltransferase alters target specificity from histidine to lysine methylation.
J.Biol.Chem., 295, 2020
|
|
