2GO0
 
 | |
1ZVN
 
 | | Crystal structure of chick MN-cadherin EC1 | | Descriptor: | Cadherin 1 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Shapiro, L, Price, S.R. | | Deposit date: | 2005-06-02 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A4E
 
 | | Crystal structure of mouse cadherin-11 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
1ZXK
 
 | | Crystal Structure of Cadherin8 EC1 domain | | Descriptor: | Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-08 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A4C
 
 | | Crystal structure of mouse cadherin-11 EC1 | | Descriptor: | Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A62
 
 | | Crystal structure of mouse cadherin-8 EC1-3 | | Descriptor: | CALCIUM ION, Cadherin-8 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Schieren, I, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-07-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
1EDP
 
 | |
6ZDH
 
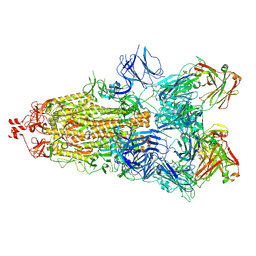 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-01 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZER
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7PQY
 
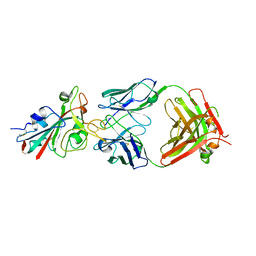 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FI-3A Fab heavy chain, FI-3A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PQZ
 
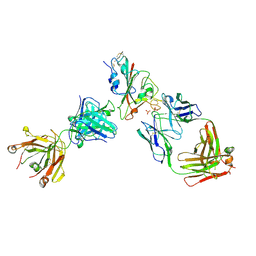 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FI-3A and FD-11A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FD-11A Fab heavy chain, FD-11A Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7PR0
 
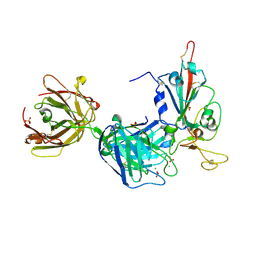 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with FD-5D Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FD-5D Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
7Q0A
 
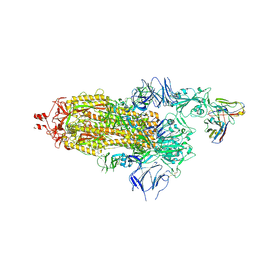 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
1QLY
 
 | | NMR Study of the SH3 Domain From Bruton's Tyrosine Kinase, 20 Structures | | Descriptor: | TYROSINE-PROTEIN KINASE BTK | | Authors: | Tzeng, S.R, Lou, Y.C, Pai, M.T, Chen, C, Chen, S.H, Cheng, J.Y. | | Deposit date: | 1999-09-20 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Btk SH3 Domain Complexed with a Proline-Rich Peptide from P120Cbl
J.Biomol.NMR, 16, 2000
|
|
8X0X
 
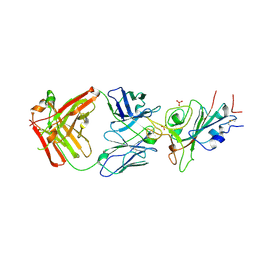 | | Crystal structure of JE-5C in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JE-5C Fab, Light chain of JE-5C Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRP
 
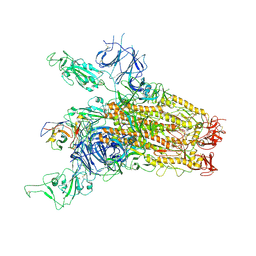 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | Descriptor: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8X0Y
 
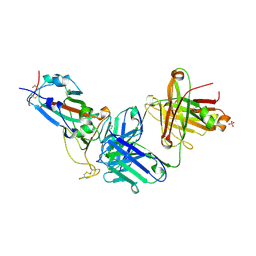 | | Crystal structure of JM-1A in complex with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of JM-1A Fab, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRO
 
 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ5
 
 | | SARS-CoV-2 Delta Spike in complex with Fab of JE-5C | | Descriptor: | Fab heavy chain of JE-5C, Fab light chain of JE-5C, Spike glycoprotein | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ6
 
 | | SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain of JH-8B, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
4JBQ
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | Aurora Kinase A, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JBO
 
 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | 1-(4-{2-[(6-{4-[2-(dimethylamino)ethoxy]phenyl}furo[2,3-d]pyrimidin-4-yl)amino]ethyl}phenyl)-3-phenylurea, Aurora kinase A | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
