5VMR
 
 | |
5VID
 
 | |
5VLI
 
 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
7SOB
 
 | |
7SOC
 
 | |
7SO9
 
 | |
7SOD
 
 | |
7SOF
 
 | |
7SOE
 
 | |
7SOA
 
 | |
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMX
 
 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7KNA
 
 | | Localized reconstruction of the H1 A/Michigan/45/2015 ectodomain displayed at the surface of I53_dn5 nanoparticle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,I53_dn5 | | Authors: | Acton, O.J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Quadrivalent influenza nanoparticle vaccines induce broad protection.
Nature, 592, 2021
|
|
6DG5
 
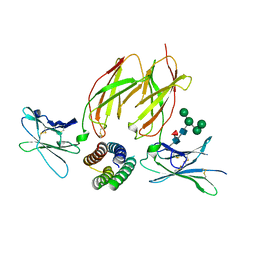 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic complex with IL-2Rb and IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.516 Å) | | Cite: | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
6DG6
 
 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic | | Descriptor: | Neoleukin-2/15 | | Authors: | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
8D06
 
 | | Hallucinated C3 protein assembly HALC3_104 | | Descriptor: | HALC3_104 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D09
 
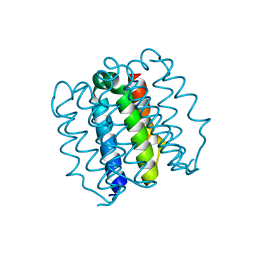 | | Hallucinated C4 protein assembly HALC4_136 | | Descriptor: | HALC4_136 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D08
 
 | | Hallucinated C4 protein assembly HALC4_135 | | Descriptor: | HALC4_135 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D04
 
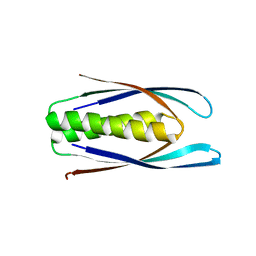 | | Hallucinated C2 protein assembly HALC2_062 | | Descriptor: | HALC2_062 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D07
 
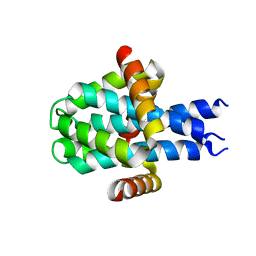 | | Hallucinated C3 protein assembly HALC3_109 | | Descriptor: | HALC3_109 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D03
 
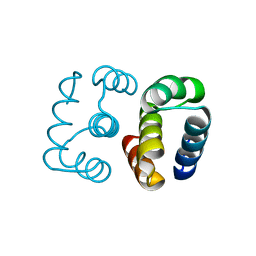 | | Hallucinated C2 protein assembly HALC2_068 | | Descriptor: | HALC2_068 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D05
 
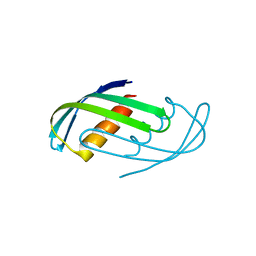 | | Hallucinated C2 protein assembly HALC2_065 | | Descriptor: | HALC2_065 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
7UR7
 
 | |
7UWZ
 
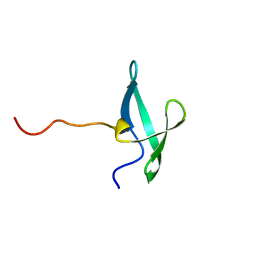 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 33_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
