1MXH
 
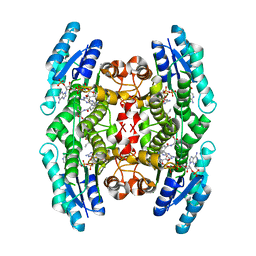 | | Crystal Structure of Substrate Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | DIHYDROFOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
1NNA
 
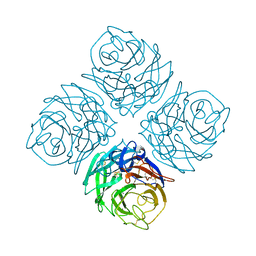 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1NNB
 
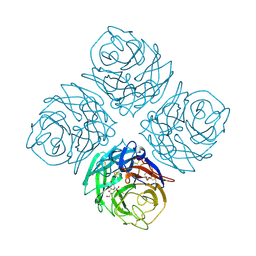 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1QCP
 
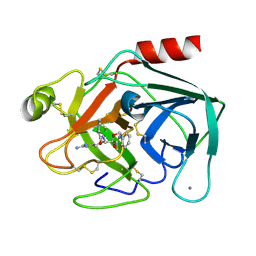 | | CRYSTAL STRUCTURE OF THE RWJ-51084 BOVINE PANCREATIC BETA-TRYPSIN AT 1.8 A | | Descriptor: | CALCIUM ION, CYCLOPENTANECARBOXYLIC ACID [1-(BENZOTHIAZOLE-2-CARBONYL)-4-GUANIDINO-BUTYL]-AMIDE, PROTEIN (BETA-TRYPSIN PROTEIN) | | Authors: | Recacha, R, Carson, M, Costanzo, M.J, Maryanoff, B, Chattopadhyay, D. | | Deposit date: | 1999-05-10 | | Release date: | 1999-05-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RWJ-51084-bovine pancreatic beta-trypsin complex at 1.8 A.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4LSJ
 
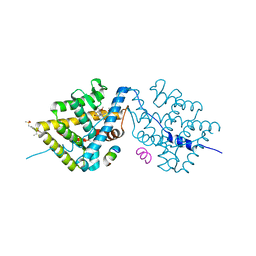 | | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Bound to a Dibenzoxapine Sulfonamide | | Descriptor: | D30 peptide, Glucocorticoid receptor, N-{3-[(1Z)-1-(10-methoxydibenzo[b,e]oxepin-11(6H)-ylidene)propyl]phenyl}methanesulfonamide | | Authors: | Carson, M, Luz, J.G, Clawson, D, Coghlan, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucocorticoid receptor modulators informed by crystallography lead to a new rationale for receptor selectivity, function, and implications for structure-based design.
J.Med.Chem., 57, 2014
|
|
1ALV
 
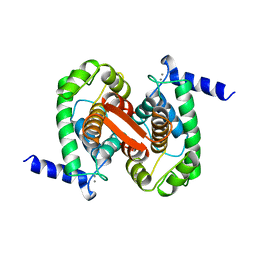 | | CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G, Chattopadhyay, D, Maki, M. | | Deposit date: | 1997-06-03 | | Release date: | 1998-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
1AMX
 
 | |
1ALW
 
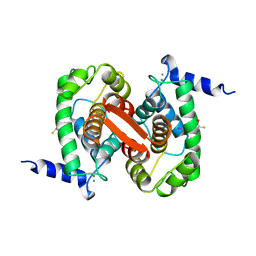 | | INHIBITOR AND CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G. | | Deposit date: | 1997-06-04 | | Release date: | 1998-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
2GP4
 
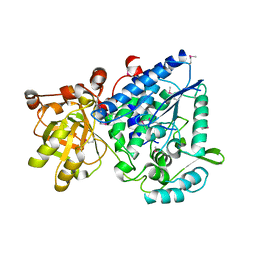 | |
1YQ1
 
 | |
1D2P
 
 | |
1D2O
 
 | |
1QC5
 
 | | I Domain from Integrin Alpha1-Beta1 | | Descriptor: | MAGNESIUM ION, PROTEIN (ALPHA1 BETA1 INTEGRIN) | | Authors: | Deivanayagam, C.C, Narayana, S.V. | | Deposit date: | 1999-05-17 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus cna MSCRAMM.
J.Biol.Chem., 274, 1999
|
|
1YO6
 
 | |
1BIO
 
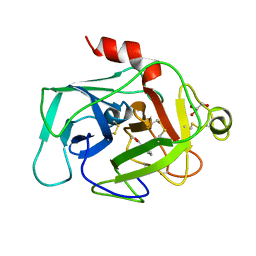 | | HUMAN COMPLEMENT FACTOR D IN COMPLEX WITH ISATOIC ANHYDRIDE INHIBITOR | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL, ISATOIC ANHYDRIDE | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
2ALR
 
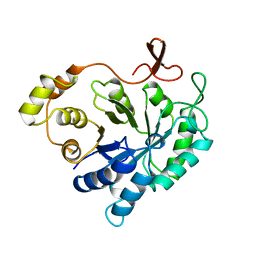 | | ALDEHYDE REDUCTASE | | Descriptor: | ALDEHYDE REDUCTASE | | Authors: | El-Kabbani, O. | | Deposit date: | 1994-09-06 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structures of human and porcine aldehyde reductase: an enzyme implicated in diabetic complications.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1DSU
 
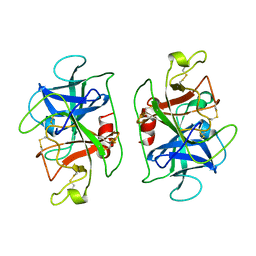 | |
1DF0
 
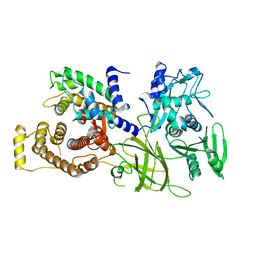 | | Crystal structure of M-Calpain | | Descriptor: | CALPAIN, M-CALPAIN | | Authors: | Hosfield, C.M, Elce, J.S, Davies, P.L, Jia, Z. | | Deposit date: | 1999-11-16 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation.
EMBO J., 18, 1999
|
|
1SN4
 
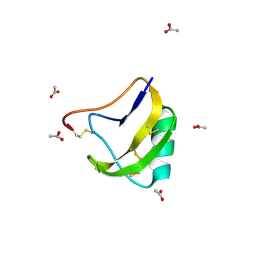 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M4 | | Descriptor: | ACETATE ION, PROTEIN (NEUROTOXIN BMK M4) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-11 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1SN1
 
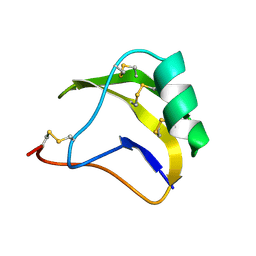 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1ROW
 
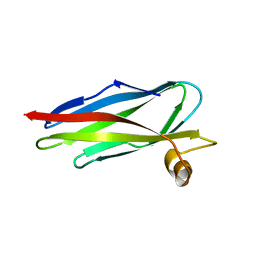 | | Structure of SSP-19, an MSP-domain protein like family member in Caenorhabditis elegans | | Descriptor: | MSP-domain protein like family member | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, J, Lu, S, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SNB
 
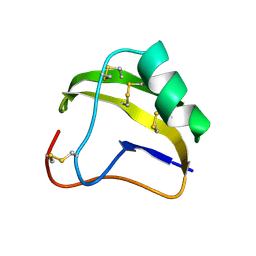 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
1DST
 
 | |
1FDP
 
 | | PROENZYME OF HUMAN COMPLEMENT FACTOR D, RECOMBINANT PROFACTOR D | | Descriptor: | PROENZYME OF COMPLEMENT FACTOR D | | Authors: | Jing, H, Macon, K.J, Moore, D, Delucas, L.J, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-12-03 | | Release date: | 1999-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D.
Embo J., 18, 1999
|
|
1HFD
 
 | | HUMAN COMPLEMENT FACTOR D IN A P21 CRYSTAL FORM | | Descriptor: | COMPLEMENT FACTOR D | | Authors: | Jing, H, Babu, Y.S, Moore, D, Kilpatrick, J.M, Liu, X.-Y, Volanakis, J.E, Narayana, S.V.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of native and complexed complement factor D: implications of the atypical His57 conformation and self-inhibitory loop in the regulation of specific serine protease activity.
J.Mol.Biol., 282, 1998
|
|
