8HDF
 
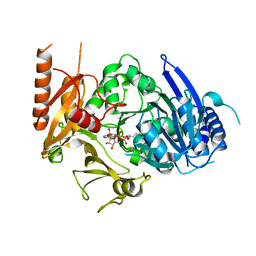 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | 分子名称: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | 登録日 | 2022-11-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
7WG6
 
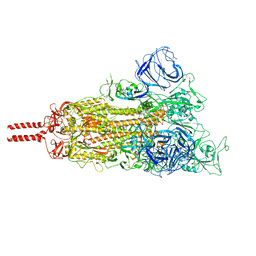 | | Neutral Omicron Spike Trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Cui, Z, Wang, X. | | 登録日 | 2021-12-28 | | 公開日 | 2022-05-18 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
8I60
 
 | |
7WGB
 
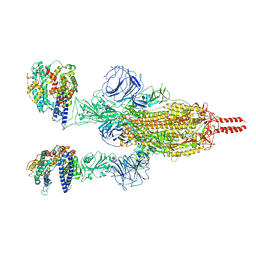 | | Neutral Omicron Spike Trimer in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WGC
 
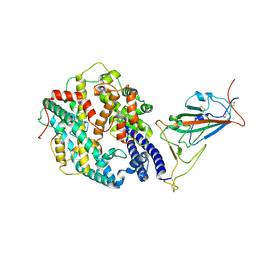 | | Neutral Omicron Spike Trimer in complex with ACE2. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 最終更新日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG7
 
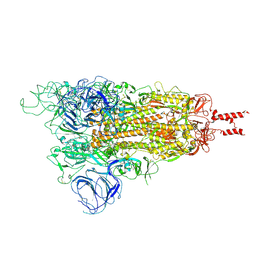 | | Acidic Omicron Spike Trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG8
 
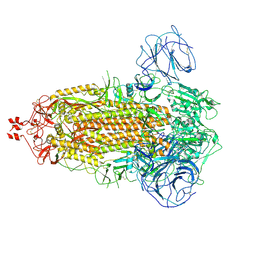 | | Delta Spike Trimer(3 RBD Down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7BQA
 
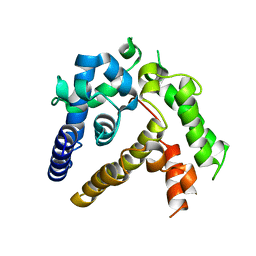 | | Crystal structure of ASFV p35 | | 分子名称: | 60 kDa polyprotein | | 著者 | Li, G.B, Fu, D, Chen, C, Guo, Y. | | 登録日 | 2020-03-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2021-05-05 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Crystal structure of the African swine fever virus structural protein p35 reveals its role for core shell assembly.
Protein Cell, 11, 2020
|
|
7WG9
 
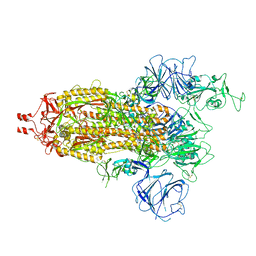 | | Delta Spike Trimer(1 RBD Up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cui, Z. | | 登録日 | 2021-12-28 | | 公開日 | 2022-06-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
6CA7
 
 | |
6CA9
 
 | |
6CA6
 
 | |
6DCQ
 
 | | Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Rantalainen, K. | | 登録日 | 2018-05-08 | | 公開日 | 2018-06-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Co-evolution of HIV Envelope and Apex-Targeting Neutralizing Antibody Lineage Provides Benchmarks for Vaccine Design.
Cell Rep, 23, 2018
|
|
8K8H
 
 | |
8P94
 
 | |
8C0F
 
 | | Tubulin-PTC596 complex | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-fluoranyl-2-(6-fluoranyl-2-methyl-benzimidazol-1-yl)-~{N}4-[4-(trifluoromethyl)phenyl]pyrimidine-4,6-diamine, ... | | 著者 | Prota, A.E, Muehlethaler, T, Weetall, M, Steinmetz, M.O. | | 登録日 | 2022-12-16 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1005 Å) | | 主引用文献 | Preclinical and Early Clinical Development of PTC596, a Novel Small-Molecule Tubulin-Binding Agent
Mol Cancer Ther, 20, 2021
|
|
5H48
 
 | | Crystal structure of Cbln1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H49
 
 | | Crystal structure of Cbln1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | 著者 | Zhong, C, Shen, J, Zhang, H, Ding, J. | | 登録日 | 2016-10-31 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5ZUD
 
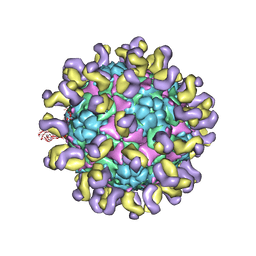 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 | | 分子名称: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | 著者 | Wang, X, Zhu, L, Wang, N. | | 登録日 | 2018-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
5ZUF
 
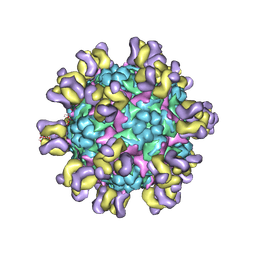 | | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with A9 | | 分子名称: | Capsid protein VP1, R10 ANTIBODY HEAVY CHAIN, R10 ANTIBODY LIGHT CHAIN, ... | | 著者 | Wang, X, Zhu, L, Wang, N. | | 登録日 | 2018-05-07 | | 公開日 | 2019-12-25 | | 最終更新日 | 2021-01-06 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71.
Mbio, 9, 2018
|
|
6IHB
 
 | |
6IH9
 
 | |
6IER
 
 | | Apo structure of a beta-glucosidase 1317 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, beta-glucosidase 1317 | | 著者 | Xie, W, Liu, X. | | 登録日 | 2018-09-16 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.246 Å) | | 主引用文献 | Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design.
Int.J.Biol.Macromol., 136, 2019
|
|
6J02
 
 | |
