4WMW
 
 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMS
 
 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMR
 
 | | STRUCTURE OF MCL1 BOUND TO BRD inhibitor ligand 1 AT 1.7A | | Descriptor: | 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, PYROPHOSPHATE 2-, ... | | Authors: | CLIFTON, M.C, EDWARDS, T.E. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMX
 
 | | The structure of MBP-MCL1 bound to ligand 6 at 2.0A | | Descriptor: | 4-ethenyl-2-[(phenylsulfonyl)amino]benzoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMV
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 4 AT 2.4A | | Descriptor: | 3-chloro-6-fluoro-1-benzothiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Moulin, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMT
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 1 AT 2.35A | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMU
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 2 AT 1.55A | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Faiman, J.W. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
3K3L
 
 | |
3CMP
 
 | |
3HWG
 
 | |
3I0A
 
 | |
3HWF
 
 | |
3HWD
 
 | |
3HWE
 
 | |
3TCQ
 
 | | Crystal Structure of matrix protein VP40 from Ebola virus Sudan | | Descriptor: | Matrix protein VP40 | | Authors: | Clifton, M.C, Edwards, T.E, Anderson, V, Atkins, K, Raymond, A, Saphire, E.O, Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution Crystal Structure of Dimeric VP40 From Sudan ebolavirus.
J Infect Dis, 212 Suppl 2, 2015
|
|
3TNY
 
 | | Structure of YfiY from Bacillus cereus bound to the siderophore iron (III) schizokinen | | Descriptor: | YfiY (ABC transport system substrate-binding protein), [2-(hydroxy-kappaO)-4-[(3-{(hydroxy-kappaO)[1-(hydroxy-kappaO)ethenyl]amino}propyl)amino]-2-{2-[(3-{(hydroxy-kappaO)[1- (hydroxy-kappaO)ethenyl]amino}propyl)amino]-2-oxoethyl}-4-oxobutanoato(6-)-kappaO]iron | | Authors: | Clifton, M.C. | | Deposit date: | 2011-09-02 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Parsing the functional specificity of Siderocalin / Lipocalin 2 / NGAL for siderophores and related small-molecule ligands
To be Published
|
|
4WGI
 
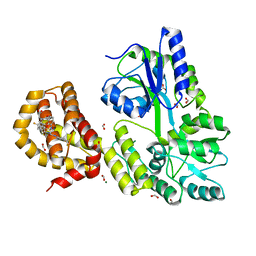 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
8U16
 
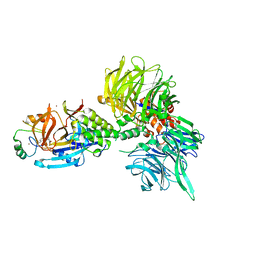 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U17
 
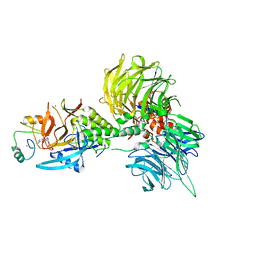 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-long bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U15
 
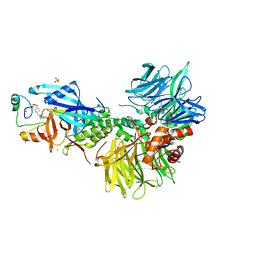 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DDB1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8TZX
 
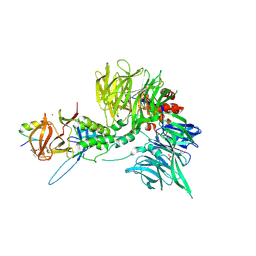 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1 | | Descriptor: | (3S)-3-(5-{(1R)-1-[(2R)-1-ethylpiperidin-2-yl]ethoxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, DNA damage-binding protein 1, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A molecular glue degrader of the WIZ transcription factor for fetal hemoglobin induction.
Science, 385, 2024
|
|
4XUZ
 
 | | Structure of CTX-M-15 bound to RPX-7009 at 1.5 A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Gardberg, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of a Cyclic Boronic Acid beta-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases.
J.Med.Chem., 58, 2015
|
|
4XUX
 
 | | Structure of ampC bound to RPX-7009 at 1.75 A | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Beta-lactamase, ... | | Authors: | Clifton, M.C, Abendroth, J. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of a Cyclic Boronic Acid beta-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases.
J.Med.Chem., 58, 2015
|
|
6O5D
 
 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
7TI2
 
 | | Structure of KPC-2 bound to RPX-7063 at 1.75A | | Descriptor: | 1,2-ETHANEDIOL, Carbapenem-hydrolyzing beta-lactamase KPC, {(3R,7S)-2-hydroxy-3-[2-(thiophen-2-yl)acetamido]-2,3,4,7-tetrahydro-1,2-oxaborepin-7-yl}acetic acid | | Authors: | Clifton, M.C, Fairman, J.W, Edwards, T.E, Hecker, S.J. | | Deposit date: | 2022-01-12 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Broad-spectrum cyclic boronate beta-lactamase inhibitors featuring an intramolecular prodrug for oral bioavailability.
Bioorg.Med.Chem., 62, 2022
|
|
