5LRX
 
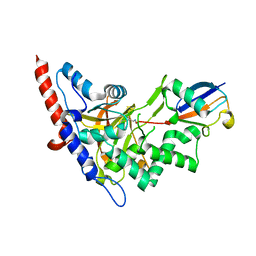 | | Structure of A20 OTU domain bound to ubiquitin | | Descriptor: | Polyubiquitin-B, Tumor necrosis factor alpha-induced protein 3 | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5TDQ
 
 | | Crystal Structure of the GOLD domain of ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | McPhail, J.A, Burke, J.E. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | The Molecular Basis of Aichi Virus 3A Protein Activation of Phosphatidylinositol 4 Kinase III beta , PI4KB, through ACBD3.
Structure, 25, 2017
|
|
5V6V
 
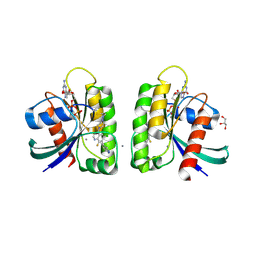 | | Crystal structure of small molecule aziridine 3 covalently bound to K-Ras G12C | | Descriptor: | 3-amino-1-{4-[6-chloro-8-fluoro-7-(5-methyl-1H-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | McGregor, L.M, Jenkins, M, Kerwin, C, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-28 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Expanding the Scope of Electrophiles Capable of Targeting K-Ras Oncogenes.
Biochemistry, 56, 2017
|
|
5VBZ
 
 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to H-Ras M72C GppNHp | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
5V6S
 
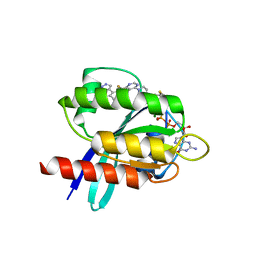 | | Crystal structure of small molecule acrylamide 1 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-8-fluoro-7-(5-methyl-1H-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | McGregor, L.M, Jenkins, M, Kerwin, C, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanding the Scope of Electrophiles Capable of Targeting K-Ras Oncogenes.
Biochemistry, 56, 2017
|
|
5VBE
 
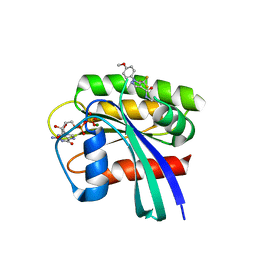 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to H-Ras M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
5VBM
 
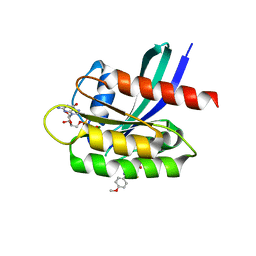 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to K-Ras Cys Light M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
4TMU
 
 | |
7APJ
 
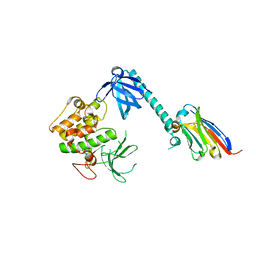 | | Structure of autoinhibited Akt1 reveals mechanism of PIP3-mediated activation | | Descriptor: | NB41, RAC-alpha serine/threonine-protein kinase,Non-specific serine/threonine protein kinase,RAC-alpha serine/threonine-protein kinase | | Authors: | Truebestein, L, Hornegger, H, Leonard, T.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of autoinhibited Akt1 reveals mechanism of PIP 3 -mediated activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6M8N
 
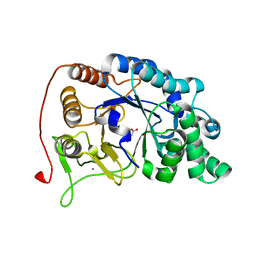 | | Endo-fucoidan hydrolase P5AFcnA from glycoside hydrolase family 107 | | Descriptor: | CALCIUM ION, MALONATE ION, P5AFcnA | | Authors: | Boraston, A.B, Vickers, C.J, Abe, K, Salama-Alber, O. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
8DEK
 
 | |
8DF2
 
 | |
7MSU
 
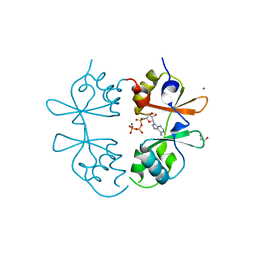 | | Crystal structure of an archaeal CNNM, MtCorB, CBS-pair domain in complex with Mg2+-ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Hemolysin, ... | | Authors: | Chen, Y.S, Gehring, K. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of an archaeal CorB magnesium transporter.
Nat Commun, 12, 2021
|
|
6TZZ
 
 | |
6TZY
 
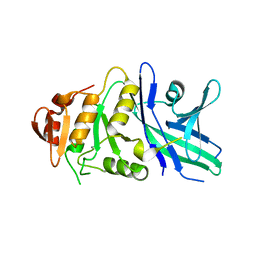 | |
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
5KC2
 
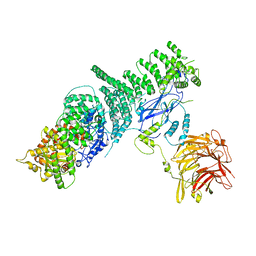 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
5KD2
 
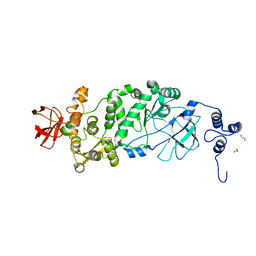 | |
5KDJ
 
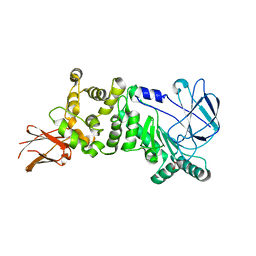 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | F5/8 type C domain protein, GLYCEROL, SODIUM ION, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDU
 
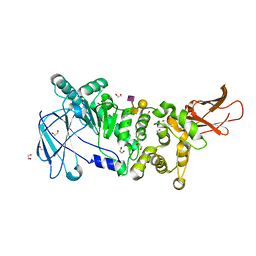 | | ZmpB metallopeptidase in complex with a2,6-Sialyl T-antigen | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, SERINE, ... | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDN
 
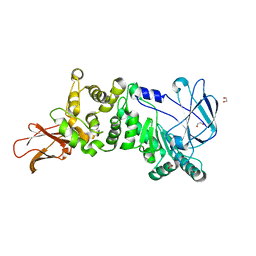 | | ZmpB metallopeptidase from Clostridium perfringens | | Descriptor: | 1,2-ETHANEDIOL, F5/8 type C domain protein, ZINC ION | | Authors: | Noach, I, Ficko-Blean, E, Stuart, C, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDX
 
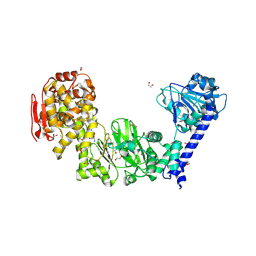 | | IMPa metallopeptidase in complex with T-antigen | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, PHOSPHATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KD8
 
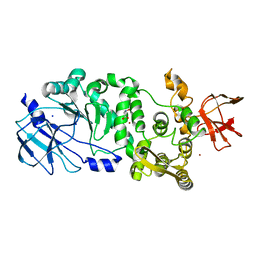 | | BT_4244 metallopeptidase in complex with Tn antigen. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Metallopeptidase, NICKEL (II) ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-07 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDV
 
 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, SULFATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDW
 
 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, PHOSPHATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
