3ZUN
 
 | | pVHL54-213-EloB-EloC complex_(2S,4R)-4-hydroxy-1-(2-(3-methylisoxazol- 5-yl)acetyl)-N-(4-nitrobenzyl)pyrrolidine-2-carboxamide bound | | Descriptor: | (4R)-N-[4-(DIHYDROXYAMINO)BENZYL]-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-L-PROLINAMIDE, GLYCEROL, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Van Molle, I, Buckley, D, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elongin-B, Elongin-C, Von Hippel-Lindau Disease Tumor Suppressor Complex
To be Published
|
|
8VLB
 
 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 4 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
8VL9
 
 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 8 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A small molecule VHL molecular glue degrader for cysteine dioxygenase 1
To Be Published
|
|
6BN8
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET55 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.990035 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BNB
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET57 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Ishoey, M, He, Z, Zhang, T, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (6.343 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN9
 
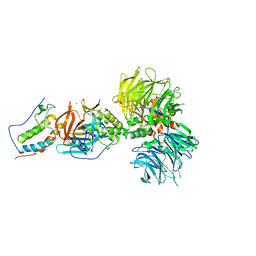 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BN7
 
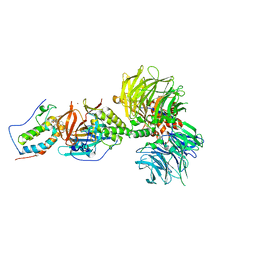 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET23 PROTAC. | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6BOY
 
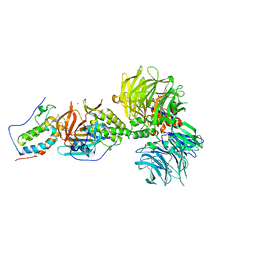 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET6 PROTAC. | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(8-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}octyl)acetamide, Bromodomain-containing protein 4, DNA damage-binding protein 1, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
2ZAL
 
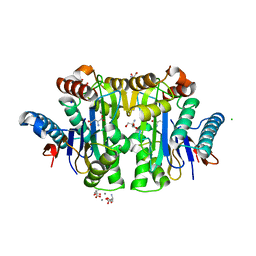 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
1K2X
 
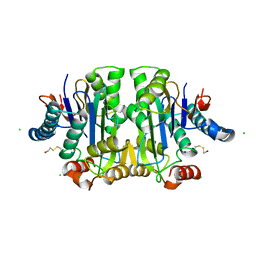 | |
1JN9
 
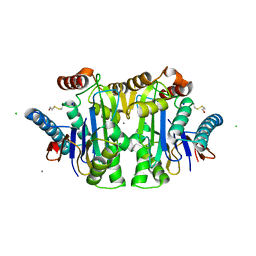 | |
2WQ9
 
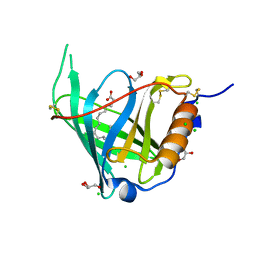 | | Crystal Structure of RBP4 bound to Oleic Acid | | Descriptor: | CHLORIDE ION, GLYCEROL, OLEIC ACID, ... | | Authors: | Nanao, M, Mercer, D, Nguyen, L, Buckley, D, Stout, T.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Rbp4 Bound to Oleic Acid
To be Published
|
|
2WQA
 
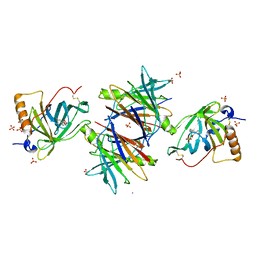 | | Complex of TTR and RBP4 and Oleic Acid | | Descriptor: | OLEIC ACID, RETINOL-BINDING PROTEIN 4, SULFATE ION, ... | | Authors: | Nanao, M, Mercer, D, Nguyen, L, Buckley, D, Stout, T.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Rbp4 Bound to Linoleic Acid and Ttr
To be Published
|
|
2GAW
 
 | |
2GAC
 
 | |
2ZAK
 
 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | Authors: | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2GEZ
 
 | | Crystal structure of potassium-independent plant asparaginase | | Descriptor: | CHLORIDE ION, L-asparaginase alpha subunit, L-asparaginase beta subunit, ... | | Authors: | Michalska, K, Bujacz, G, Jaskolski, M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant asparaginase.
J.Mol.Biol., 360, 2006
|
|
9GAC
 
 | |
9GAA
 
 | |
9GAF
 
 | |
3C17
 
 | |
