1MF2
 
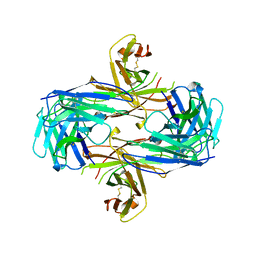 | | ANTI HIV1 PROTEASE FAB COMPLEX | | Descriptor: | MONOCLONAL ANTIBODY F11.2.32 | | Authors: | Lescar, J, Bentley, G.A. | | Deposit date: | 1996-12-27 | | Release date: | 1997-12-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of an Fab-peptide complex: structural basis of HIV-1 protease inhibition by a monoclonal antibody.
J.Mol.Biol., 267, 1997
|
|
6I1A
 
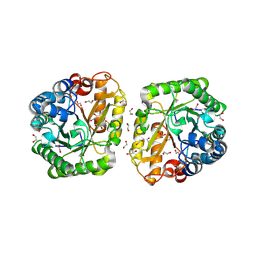 | | Crystal structure of rutinosidase from Aspergillus niger | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, rutinosidase | | Authors: | Pachl, P, Rezacova, P, Kapesova, J. | | Deposit date: | 2018-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rutinosidase from Aspergillus niger: crystal structure and insight into the enzymatic activity.
Febs J., 287, 2020
|
|
1CL7
 
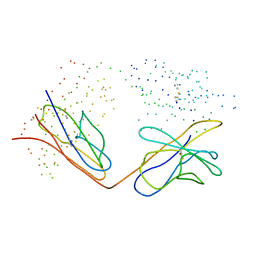 | | ANTI HIV1 PROTEASE FAB | | Descriptor: | PROTEIN (IGG1 ANTIBODY 1696 (constant heavy chain)), PROTEIN (IGG1 ANTIBODY 1696 (light chain)), PROTEIN (IGG1 ANTIBODY 1696 (variable heavy chain)) | | Authors: | Lescar, J, Bentley, G.A. | | Deposit date: | 1999-05-06 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of the HIV-1 and HIV-2 proteases by a monoclonal antibody.
Protein Sci., 8, 1999
|
|
5K57
 
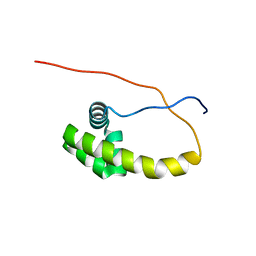 | | HDD domain from human Ddi2 | | Descriptor: | Protein DDI1 homolog 2 | | Authors: | Veverka, V. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Human DNA-Damage-Inducible 2 Protein Is Structurally and Functionally Distinct from Its Yeast Ortholog.
Sci Rep, 6, 2016
|
|
1M0B
 
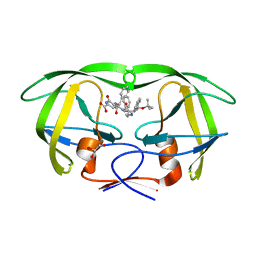 | | HIV-1 protease in complex with an ethyleneamine inhibitor | | Descriptor: | GLYCEROL, N-{(3S)-3-[(tert-butoxycarbonyl)amino]-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Petrokova, H, Hasek, J, Dohnalek, J. | | Deposit date: | 2002-06-12 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of hydroxyl group and R/S configuration of isostere in binding properties of HIV-1 protease inhibitors
Eur.J.Biochem., 271, 2004
|
|
7QBL
 
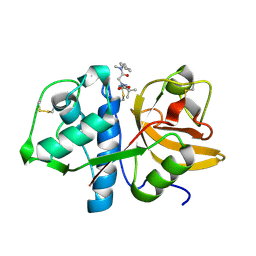 | |
7QBM
 
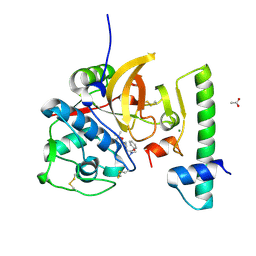 | | Structure of the activation intermediate of cathepsin K in complex with the 3-cyano-3-aza-beta-amino acid inhibitor Gu2602 | | Descriptor: | ACETATE ION, Cathepsin K, MAGNESIUM ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBN
 
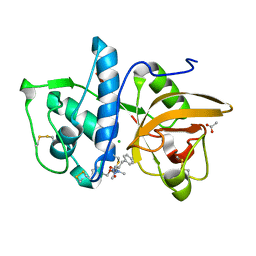 | | Structure of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7QBO
 
 | | Structure of the activation intermediate of cathepsin K in complex with the azadipeptide nitrile inhibitor Gu1303 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[aminomethyl(methyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Benysek, J, Busa, M, Mares, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
J Enzyme Inhib Med Chem, 37, 2022
|
|
3I8W
 
 | |
1W8P
 
 | | Structural properties of the B25Tyr-NMe-B26Phe insulin mutant. | | Descriptor: | INSULIN A-CHAIN, INSULIN B-CHAIN, PHENOL, ... | | Authors: | Zakowa, L, Au-Alvarez, O, Dodson, E.J, Dodson, G.G, Brzozowski, A.M. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Towards the Insulin-Igf-I Intermediate Structures: Functional and Structural Properties of the B25Tyr-Nme-B26Phe Insulin Mutant.
Biochemistry, 43, 2004
|
|
6GSZ
 
 | | Crystal structure of native alfa-L-rhamnosidase from Aspergillus terreus | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Pachl, P, Rezacova, P, Skerlova, J. | | Deposit date: | 2018-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of native alpha-L-rhamnosidase from Aspergillus terreus.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2N7D
 
 | |
7ZSL
 
 | | human purine nucleoside phosphorylase in complex with JS-196 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Purine nucleoside phosphorylase, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSN
 
 | | human purine nucleoside phosphorylase in complex with JS-379 | | Descriptor: | 6-tungstotellurate(VI), Purine nucleoside phosphorylase, SULFATE ION, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSQ
 
 | | human purine nucleoside phosphorylase in complex with JS-555 | | Descriptor: | 1,2-ETHANEDIOL, 6-tungstotellurate(VI), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSP
 
 | | human purine nucleoside phosphorylase in complex with JS-555 | | Descriptor: | 6-tungstotellurate(VI), Purine nucleoside phosphorylase, SODIUM ION, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSM
 
 | | human purine nucleoside phosphorylase in complex with JS-375 | | Descriptor: | CHLORIDE ION, Purine nucleoside phosphorylase, SULFATE ION, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSO
 
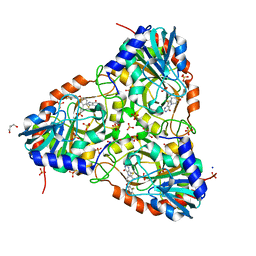 | | human purine nucleoside phosphorylase in complex with JS-554 | | Descriptor: | GLYCEROL, Purine nucleoside phosphorylase, SODIUM ION, ... | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
7ZSR
 
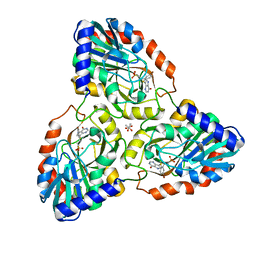 | | purine nucleoside phosphorylase in complex with JS-379 | | Descriptor: | ACETATE ION, Purine nucleoside phosphorylase, [(~{E})-2-[5-bromanyl-2-[(4-oxidanylidene-3,5-dihydropyrrolo[3,2-d]pyrimidin-7-yl)sulfanyl]phenyl]ethenyl]phosphonic acid | | Authors: | Djukic, S, Pachl, P, Rezacova, P. | | Deposit date: | 2022-05-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and Crystallographic Study of Novel Purine Nucleoside Phosphorylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
5MRK
 
 | |
6S27
 
 | |
4MUM
 
 | |
7OJ4
 
 | | Crystal structure of apo NS3 helicase from tick-borne encephalitis virus | | Descriptor: | NS3 helicase domain | | Authors: | Anindita, P.D, Havlickova, P, Kascakova, B, Grinkevich, P, Brynda, J, Franta, Z. | | Deposit date: | 2021-05-13 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
7OYO
 
 | | Carbonic anhydrase II in complex with Hit4 (MH70) | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ZINC ION, ... | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2021-06-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Identification of specific carbonic anhydrase inhibitors via in situ click chemistry, phage-display and synthetic peptide libraries: comparison of the methods and structural study.
Rsc Med Chem, 14, 2023
|
|
