7RY3
 
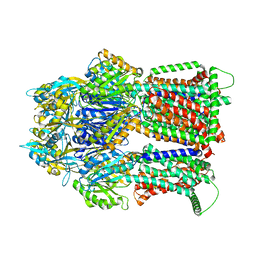 | | Multidrug Efflux pump AdeJ with TP-6076 bound | | Descriptor: | (4S,4aS,5aR,12aS)-4-(diethylamino)-3,10,12,12a-tetrahydroxy-1,11-dioxo-8-[(2S)-pyrrolidin-2-yl]-7-(trifluoromethyl)-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide, Efflux pump membrane transporter | | Authors: | Zhang, Z. | | Deposit date: | 2021-08-24 | | Release date: | 2022-02-02 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | An Analysis of the Novel Fluorocycline TP-6076 Bound to Both the Ribosome and Multidrug Efflux Pump AdeJ from Acinetobacter baumannii.
Mbio, 13, 2022
|
|
8FFS
 
 | |
8FFK
 
 | |
7TB7
 
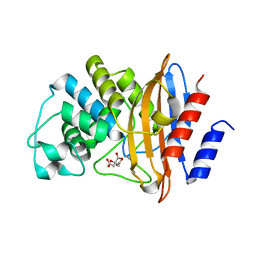 | | Crystal structure of D179N KPC-2 beta-lactamase | | Descriptor: | CITRIC ACID, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | van den Akker, F, Alsenani, T. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Characterization of the D179N and D179Y Variants of KPC-2 beta-Lactamase: Omega-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam.
Antimicrob.Agents Chemother., 66, 2022
|
|
7TBX
 
 | | Crystal structure of D179Y KPC-2 beta-lactamase | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | van den Akker, F, Alsenani, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Structural Characterization of the D179N and D179Y Variants of KPC-2 beta-Lactamase: Omega-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam.
Antimicrob.Agents Chemother., 66, 2022
|
|
7TC1
 
 | |
8G2R
 
 | | CRYSTAL STRUCTURE OF THE KPC-2 D179N VARIANT IN COMPLEX WITH AVIBACTAM | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Alsenani, T, van den Akker, F. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Exploring avibactam and relebactam inhibition of Klebsiella pneumoniae carbapenemase D179N variant: role of the Omega loop-held deacylation water.
Antimicrob.Agents Chemother., 67, 2023
|
|
8G2T
 
 | |
7LLB
 
 | | Crystal structure of KPC-2 S70G/T215P mutant with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LJK
 
 | | Crystal structure of the deacylation deficient KPC-2 F72Y mutant | | Descriptor: | Beta-lactamase | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-01-29 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
7LLH
 
 | | KPC-2 F72Y mutant with acylated imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Furey, I, Palzkill, T, Sankaran, B, Hu, L, Prasad, B.V.V. | | Deposit date: | 2021-02-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Local interactions with the Glu166 base and the conformation of an active site loop play key roles in carbapenem hydrolysis by the KPC-2 beta-lactamase.
J.Biol.Chem., 296, 2021
|
|
