1NU7
 
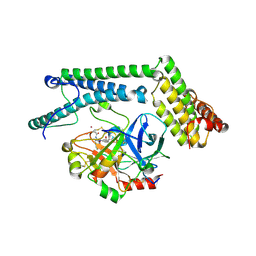 | | Staphylocoagulase-Thrombin Complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
1NU9
 
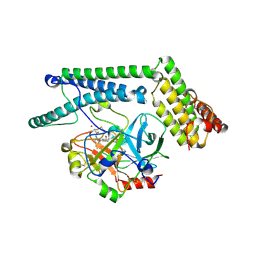 | | Staphylocoagulase-Prethrombin-2 complex | | Descriptor: | IMIDAZOLE, MERCURY (II) ION, N-(sulfanylacetyl)-D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Friedrich, R, Bode, W, Fuentes-Prior, P, Panizzi, P, Bock, P.E. | | Deposit date: | 2003-01-31 | | Release date: | 2003-10-07 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation
NATURE, 425, 2003
|
|
1OP8
 
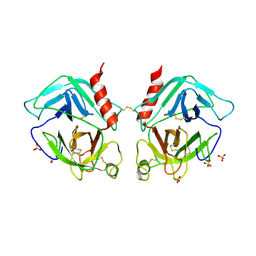 | | Crystal Structure of Human Granzyme A | | Descriptor: | Granzyme A, SULFATE ION | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Bode, W, Jenne, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the apoptosis-inducing human granzyme A dimer
NAT.STRUCT.BIOL., 10, 2003
|
|
1ORW
 
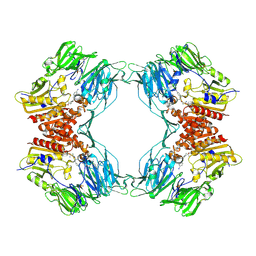 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | Descriptor: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P8J
 
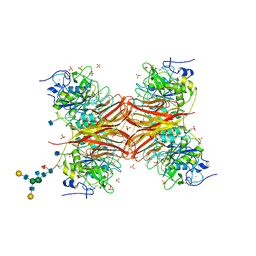 | | CRYSTAL STRUCTURE OF THE PROPROTEIN CONVERTASE FURIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Henrich, S, Cameron, A, Bourenkov, G.P, Kiefersauer, R, Huber, R, Lindberg, I, Bode, W, Than, M.E. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Proprotein Processing Proteinase Furin Explains its Stringent Specificity
Nat.Struct.Biol., 10, 2003
|
|
1ORV
 
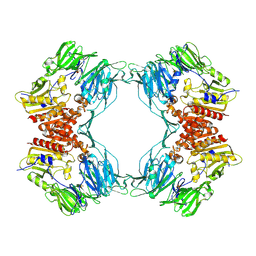 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1PEX
 
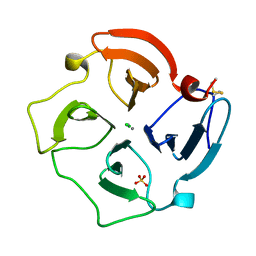 | | COLLAGENASE-3 (MMP-13) C-TERMINAL HEMOPEXIN-LIKE DOMAIN | | Descriptor: | CALCIUM ION, CHLORIDE ION, COLLAGENASE-3, ... | | Authors: | Gomis-Ruth, F.X, Gohlke, U, Betz, M, Knauper, V, Murphy, G, Lopez-Otin, C, Bode, W. | | Deposit date: | 1996-05-24 | | Release date: | 1996-12-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The helping hand of collagenase-3 (MMP-13): 2.7 A crystal structure of its C-terminal haemopexin-like domain.
J.Mol.Biol., 264, 1996
|
|
1PFX
 
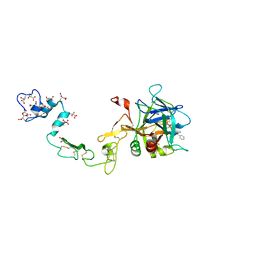 | | PORCINE FACTOR IXA | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, FACTOR IXA | | Authors: | Brandstetter, H, Bauer, M, Huber, R, Lollar, P, Bode, W. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of clotting factor IXa: active site and module structure related to Xase activity and hemophilia B.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1PJP
 
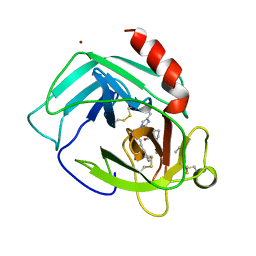 | | THE 2.2 A CRYSTAL STRUCTURE OF HUMAN CHYMASE IN COMPLEX WITH SUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYLKETONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase, SUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Pereira, P.J.B, Wang, Z.M, Rubin, H, Huber, R, Bode, W, Schechter, N.M, Strobl, S. | | Deposit date: | 1998-09-07 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A crystal structure of human chymase in complex with succinyl-Ala-Ala-Pro-Phe-chloromethylketone: structural explanation for its dipeptidyl carboxypeptidase specificity.
J.Mol.Biol., 286, 1999
|
|
1POJ
 
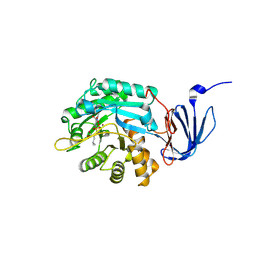 | | Isoaspartyl Dipeptidase with bound inhibitor | | Descriptor: | 2-{[[(1S)-1-AMINO-2-CARBOXYETHYL](DIHYDROXY)PHOSPHORANYL]METHYL}-4-METHYLPENTANOIC ACID, Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1POK
 
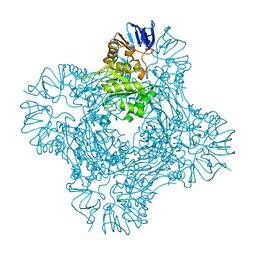 | | Crystal structure of Isoaspartyl Dipeptidase | | Descriptor: | ASPARAGINE, Isoaspartyl dipeptidase, SULFATE ION, ... | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1PO9
 
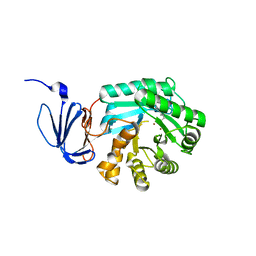 | | Crytsal structure of isoaspartyl dipeptidase | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1RM8
 
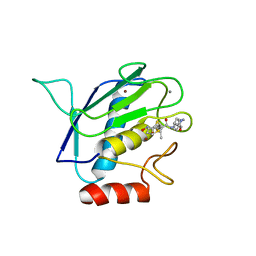 | | Crystal structure of the catalytic domain of MMP-16/MT3-MMP: Characterization of MT-MMP specific features | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, Matrix metalloproteinase-16, ... | | Authors: | Lang, R, Braun, M, Sounni, N.E, Noel, A, Frankenne, F, Foidart, J.-M, Bode, W, Maskos, K. | | Deposit date: | 2003-11-27 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the catalytic domain of MMP-16/MT3-MMP: characterization of MT-MMP specific features.
J.Mol.Biol., 336, 2004
|
|
1RFN
 
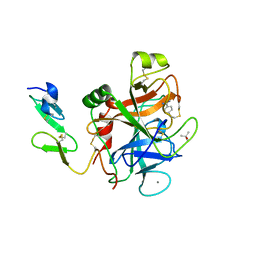 | | HUMAN COAGULATION FACTOR IXA IN COMPLEX WITH P-AMINO BENZAMIDINE | | Descriptor: | CALCIUM ION, P-AMINO BENZAMIDINE, PROTEIN (COAGULATION FACTOR IX), ... | | Authors: | Hopfner, K.-P, Lang, A, Karcher, A, Sichler, K, Kopetzki, E, Brandstetter, H, Huber, R, Bode, W, Engh, R.A. | | Deposit date: | 1999-04-19 | | Release date: | 1999-09-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding.
Structure Fold.Des., 7, 1999
|
|
1GTJ
 
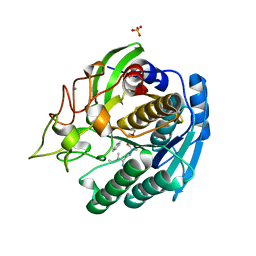 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolisin (KSCP) - complex with Ac-Ile-Ala-Phe-cho | | Descriptor: | ALDEHYDE INHIBITOR, CALCIUM ION, KUMAMOLYSIN, ... | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1GTG
 
 | | Crystal structure of the thermostable serine-carboxyl type proteinase, kumamolysin (kscp) | | Descriptor: | CALCIUM ION, KUMAMOLYSIN | | Authors: | Comellas-Bigler, M, Fuentes-Prior, P, Maskos, K, Huber, R, Oyama, H, Uchida, K, Dunn, B.M, Oda, K, Bode, W. | | Deposit date: | 2002-01-15 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.4 A Crystal Structure of Kumamolysin. A Thermostable Serine-Carboxyl-Type Proteinase
Structure, 10, 2002
|
|
1HLE
 
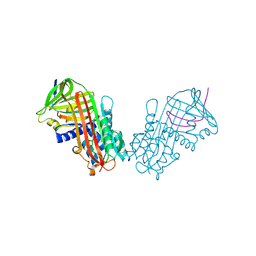 | | CRYSTAL STRUCTURE OF CLEAVED EQUINE LEUCOCYTE ELASTASE INHIBITOR DETERMINED AT 1.95 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, HORSE LEUKOCYTE ELASTASE INHIBITOR | | Authors: | Baumann, U, Bode, W, Huber, R, Travis, J, Potempa, J. | | Deposit date: | 1992-04-13 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
J.Mol.Biol., 226, 1992
|
|
1HTD
 
 | | STRUCTURAL INTERACTION OF NATURAL AND SYNTHETIC INHIBITORS WITH THE VENOM METALLOPROTEINASE, ATROLYSIN C (HT-D) | | Descriptor: | ATROLYSIN C, CALCIUM ION, ZINC ION | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1994-01-20 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1HUC
 
 | |
1JC9
 
 | | TACHYLECTIN 5A FROM TACHYPLEUS TRIDENTATUS (JAPANESE HORSESHOE CRAB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, techylectin-5A | | Authors: | Kairies, N, Beisel, H.-G, Fuentes-Prior, P, Tsuda, R, Muta, T, Iwanaga, S, Bode, W, Huber, R, Kawabata, S. | | Deposit date: | 2001-06-08 | | Release date: | 2001-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JQG
 
 | | Crystal Structure of the Carboxypeptidase A from Helicoverpa Armigera | | Descriptor: | ZINC ION, carboxypeptidase A | | Authors: | Estebanez-Perpina, E, Bayes, A, Vendrell, J, Jongsma, M.A, Bown, D.P, Gatehouse, J.A, Huber, R, Bode, W, Aviles, F.X, Reverter, D. | | Deposit date: | 2001-08-07 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a novel mid-gut procarboxypeptidase from the cotton pest Helicoverpa armigera.
J.Mol.Biol., 313, 2001
|
|
1JK3
 
 | | Crystal structure of human MMP-12 (Macrophage Elastase) at true atomic resolution | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, MACROPHAGE METALLOELASTASE, ... | | Authors: | Lang, R, Kocourek, A, Braun, M, Tschesche, H, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2001-07-11 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Substrate specificity determinants of human macrophage elastase (MMP-12) based on the 1.1 A crystal structure.
J.Mol.Biol., 312, 2001
|
|
1KBC
 
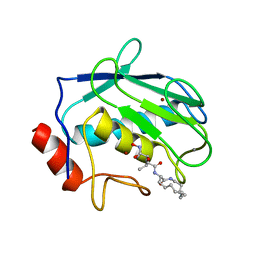 | | PROCARBOXYPEPTIDASE TERNARY COMPLEX | | Descriptor: | 3-AMINO-AZACYCLOTRIDECAN-2-ONE, 3-FORMYL-2-HYDROXY-5-METHYL-HEXANOIC ACID HYDROXYAMIDE, CALCIUM ION, ... | | Authors: | Betz, M, Gomis-Rueth, F.X, Bode, W. | | Deposit date: | 1997-04-29 | | Release date: | 1997-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8-A crystal structure of the catalytic domain of human neutrophil collagenase (matrix metalloproteinase-8) complexed with a peptidomimetic hydroxamate primed-side inhibitor with a distinct selectivity profile.
Eur.J.Biochem., 247, 1997
|
|
1KLI
 
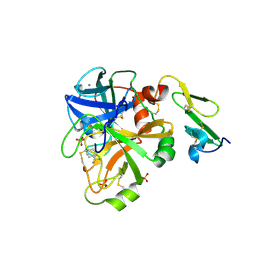 | | Cofactor-and substrate-assisted activation of factor VIIa | | Descriptor: | BENZAMIDINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Sichler, K, Banner, D.W, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of Uninhibited Factor VIIa Link its Cofactor and Substrate-assisted Activation to Specific Interactions
J.Mol.Biol., 322, 2002
|
|
1KLJ
 
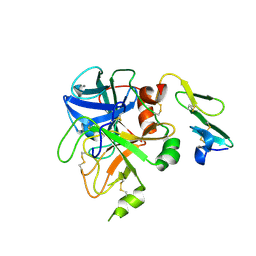 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
