8SEL
 
 | |
3IPV
 
 | | Crystal structure of Spatholobus parviflorus seed lectin | | Descriptor: | CALCIUM ION, Lectin alpha chain, Lectin beta chain, ... | | Authors: | Geethanandan, K, Bharath, S.R, Abhilash, J, Sadasivan, C, Haridas, M. | | Deposit date: | 2009-08-18 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | X-ray structure of a galactose-specific lectin from Spatholobous parviflorous
Int.J.Biol.Macromol., 49, 2011
|
|
7X0F
 
 | |
7X0E
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | Descriptor: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
7X17
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC bound to Ppant-L-Ala | | Descriptor: | AMB antimetabolite synthase AmbB, S-[2-[3-[[(2S)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (2R)-2-azanylpropanethioate | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
7TMC
 
 | |
7UN5
 
 | |
7UMQ
 
 | |
5XX1
 
 | | Crystal structure of Arginine decarboxylase (AdiA) from Salmonella typhimurium | | Descriptor: | Arginine decarboxylase, PHOSPHATE ION | | Authors: | Deka, G, Bharath, S.R, Shavithri, H.S, Murthy, M.R.N. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural studies on the decameric S. typhimurium arginine decarboxylase (ADC): Pyridoxal 5'-phosphate binding induces conformational changes
Biochem. Biophys. Res. Commun., 490, 2017
|
|
7BV7
 
 | | INTS3 complexed with INTS6 | | Descriptor: | Integrator complex subunit 3, Integrator complex subunit 6 | | Authors: | Jia, Y, Bharath, S.R, Song, H. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-14 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair.
Cell Discov, 7, 2021
|
|
5ZMC
 
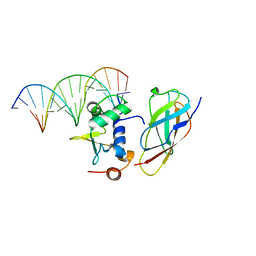 | | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*GP*AP*CP*CP*CP*GP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*GP*CP*CP*CP*TP*TP*CP*CP*GP*GP*GP*TP*CP*CP*CP*C)-3'), Nuclear factor NF-kappa-B p100 subunit, ... | | Authors: | Xu, X, Bharath, S.R, Song, H. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Nat Commun, 9, 2018
|
|
6AA9
 
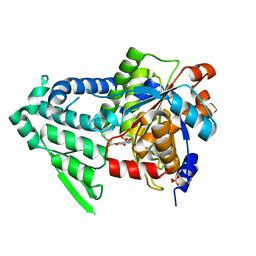 | | T166A mutant of D-Serine deaminase from Salmonella typhimurium | | Descriptor: | D-serine dehydratase, PHOSPHATE ION, SODIUM ION | | Authors: | Deka, G, Bharath, S.R, Shavithri, H.S, Murthy, M.R.N. | | Deposit date: | 2018-07-17 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on the decameric S. typhimurium arginine decarboxylase (ADC): Pyridoxal 5'-phosphate binding induces conformational changes
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5FHD
 
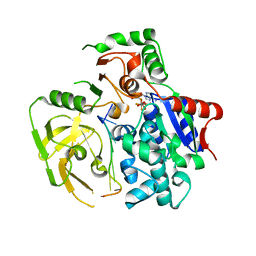 | | Structure of Bacteroides sp Pif1 complexed with tailed dsDNA resulting in ssDNA bound complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*CP*GP*GP*GP*GP*CP*CP*GP*CP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FHH
 
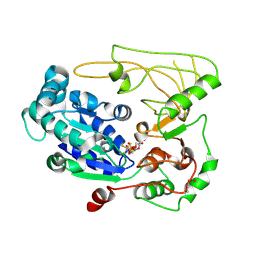 | | Structure of human Pif1 helicase domain residues 200-641 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, TETRAFLUOROALUMINATE ION | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FHG
 
 | |
5FHF
 
 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5FHE
 
 | | Crystal structure of Bacteroides Pif1 bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
4JEX
 
 | | Y21K mutant of N-acetylornithine aminotransferase complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4JEZ
 
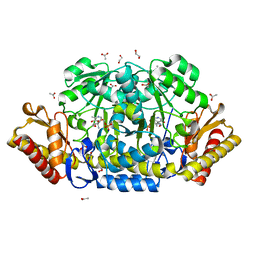 | | N79R mutant of N-acetylornithine aminotransferase complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4JEV
 
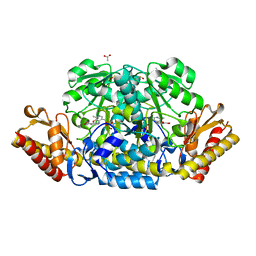 | | N-acetylornithine aminotransferase from S. typhimurium complexed with gabaculine | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases.
To be Published
|
|
4JF1
 
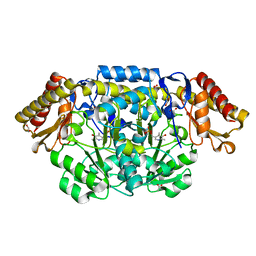 | | R144Q mutant of N-acetylornithine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4JEW
 
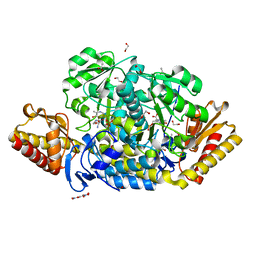 | | N-acetylornithine aminotransferase from S. typhimurium complexed with L-canaline | | Descriptor: | (2S)-2-azanyl-4-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxy-butanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases.
To be Published
|
|
4JF0
 
 | | N79R mutant of N-acetylornithine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4JEY
 
 | | E198A mutant of N-acetylornithine aminotransferase from Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Acetylornithine/succinyldiaminopimelate aminotransferase, ... | | Authors: | Bisht, S, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational transitions, ligand specificity and catalysis in N-acetylornithine aminotransferase: Implications on drug designing and rational enzyme engineering in omega aminotransferases
To be Published
|
|
4D9I
 
 | | Crystal structure of holo Diaminopropionate ammonia lyase from Escherichia coli | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diaminopropionate ammonia-lyase | | Authors: | Bisht, S, Rajaram, V, Bharath, S.R, Murthy, M.R.N. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Escherichia coli Diaminopropionate Ammonia-lyase Reveals Mechanism of Enzyme Activation and Catalysis
J.Biol.Chem., 287, 2012
|
|
