4FF6
 
 | | Mycobacterium tuberculosis DprE1 in complex with CT325 - monoclinic crystal form | | Descriptor: | 3-(hydroxyamino)-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FDN
 
 | |
4FDP
 
 | |
4FDO
 
 | | Mycobacterium tuberculosis DprE1 in complex with CT319 | | Descriptor: | 3-nitro-N-[(1R)-1-phenylethyl]-5-(trifluoromethyl)benzamide, FLAVIN-ADENINE DINUCLEOTIDE, oxidoreductase DprE1 | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2012-05-29 | | Release date: | 2012-07-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3FVE
 
 | | Crystal structure of diaminopimelate epimerase Mycobacterium tuberculosis DapF | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Diaminopimelate epimerase, GLYCEROL | | Authors: | Usha, V, Dover, L.G, Roper, D.I, Futterer, K, Besra, G.S. | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5DU8
 
 | | Crystal structure of M. tuberculosis EchA6 bound to GSK572A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-[(5-fluoropyridin-2-yl)methyl]-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DTP
 
 | |
5DU4
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK366A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-N-(4-methoxybenzyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DTW
 
 | |
5DUC
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK951A | | Descriptor: | (5R,7S)-N-(1,3-benzodioxol-5-ylmethyl)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DUF
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK729A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxylic acid, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DU6
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK059A. | | Descriptor: | (5R,7R)-5-(4-ethylphenyl)-N-(4-fluorobenzyl)-7-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
3PTY
 
 | | Crystal structure of the C-terminal extracellular domain of Mycobacterium tuberculosis EmbC | | Descriptor: | Arabinosyltransferase C, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Alderwick, L.J, Besra, G.S, Futterer, K. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The C-Terminal Domain of the Arabinosyltransferase Mycobacterium tuberculosis EmbC Is a Lectin-Like Carbohydrate Binding Module.
Plos Pathog., 7, 2011
|
|
2Q74
 
 | | Mycobacterium tuberculosis SuhB | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Brown, A.K, Meng, G, Ghadbane, H, Besra, G.S, Futterer, K. | | Deposit date: | 2007-06-06 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+
Bmc Struct.Biol., 7, 2007
|
|
2QJ3
 
 | | Mycobacterium tuberculosis FabD | | Descriptor: | Malonyl CoA-acyl carrier protein transacylase, NICKEL (II) ION | | Authors: | Ghadbane, H, Brown, A.K, Kremer, L, Besra, G.S, Futterer, K. | | Deposit date: | 2007-07-06 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mycobacterium tuberculosis mtFabD, a malonyl-CoA:acyl carrier protein transacylase (MCAT).
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2FEZ
 
 | | Mycobacterium tuberculosis EmbR | | Descriptor: | Probable regulatory protein embR | | Authors: | Futterer, K, Alderwick, L.J, Besra, G.S. | | Deposit date: | 2005-12-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FF4
 
 | |
3OKA
 
 | | Crystal structure of Corynebacterium glutamicum PimB' in complex with GDP-Man (triclinic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
3OKC
 
 | | Crystal structure of Corynebacterium glutamicum PimB' bound to GDP (orthorhombic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
3OKP
 
 | | Crystal structure of Corynebacterium glutamicum PimB' bound to GDP-Man (orthorhombic crystal form) | | Descriptor: | GDP-mannose-dependent alpha-(1-6)-phosphatidylinositol monomannoside mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE | | Authors: | Batt, S.M, Jabeen, T, Besra, G.S, Futterer, K. | | Deposit date: | 2010-08-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acceptor substrate discrimination in phosphatidyl-myo-inositol mannoside synthesis: structural and mutational analysis of mannosyltransferase Corynebacterium glutamicum PimB'.
J.Biol.Chem., 285, 2010
|
|
7UFJ
 
 | | Structure of human MR1-ethylvanillin in complex with human MAIT A-F7 TCR | | Descriptor: | 3-ethoxy-4-hydroxybenzaldehyde, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wang, C.J, Rossjohn, J, Le Nours, J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative affinity measurement of small molecule ligand binding to Major Histocompatibility Complex class-I related protein 1 MR1.
J.Biol.Chem., 298, 2022
|
|
7BX8
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | Descriptor: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7BWR
 
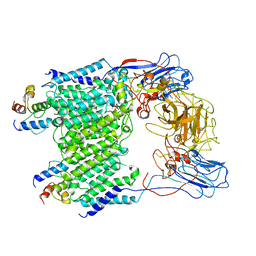 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
5W0C
 
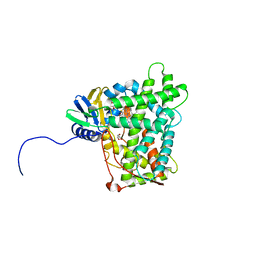 | | Cytochrome P450 (CYP) 2C9 TCA007 Inhibitor Complex | | Descriptor: | Cytochrome P450 2C9, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnson, E.F, Hsu, M.-H. | | Deposit date: | 2017-05-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7KP0
 
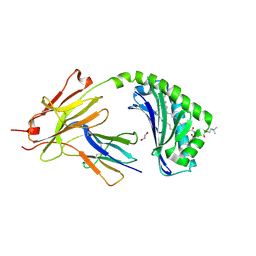 | | CD1a-42:1 SM binary complex | | Descriptor: | (4R,7S)-4-hydroxy-7-[(1S,2E)-1-hydroxyhexadec-2-en-1-yl]-N,N,N-trimethyl-4,9-dioxo-3,5-dioxa-8-aza-4lambda~5~-phosphadotriacontan-1-aminium, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Wegrecki, M, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-11-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD1a selectively captures endogenous cellular lipids that broadly block T cell response.
J.Exp.Med., 218, 2021
|
|
