2M5X
 
 | | Novel method of protein purification for structural research. Example of ultra high resolution structure of SPI-2 inhibitor by X-ray and NMR spectroscopy. | | Descriptor: | Silk protease inhibitor 2 | | Authors: | Lenarcic Zivkovic, M, Dvornyk, A, Kludkiewicz, B, Kopera, E, Zagorski-Ostoja, W, Grzelak, K, Zhukov, I, Bal, W. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
4HGU
 
 | | Crystal Structure of Galleria mellonella Silk Protease Inhibitor 2 | | Descriptor: | SODIUM ION, Silk protease inhibitor 2 | | Authors: | Krzywda, S, Jaskolski, M, Dvornyk, A, Kludkiewicz, B, Grzelak, K, Zagorski, W, Bal, W, Kopera, E. | | Deposit date: | 2012-10-08 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
1IAR
 
 | | INTERLEUKIN-4 / RECEPTOR ALPHA CHAIN COMPLEX | | Descriptor: | PROTEIN (INTERLEUKIN-4 RECEPTOR ALPHA CHAIN), PROTEIN (INTERLEUKIN-4) | | Authors: | Hage, T, Sebald, W, Reinemer, P. | | Deposit date: | 1999-02-25 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the interleukin-4/receptor alpha chain complex reveals a mosaic binding interface.
Cell(Cambridge,Mass.), 97, 1999
|
|
5N4K
 
 | | N-terminal domain of a human Coronavirus NL63 nucleocapsid protein | | Descriptor: | Nucleoprotein, SULFATE ION | | Authors: | Zdzalik, M, Szelazek, B, Kabala, W, Golik, P, Burmistrz, M, Florek, D, Kus, K, Pyrc, K, Dubin, G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
8CH7
 
 | | RDC-refined Interleukin-4 (wild type) pH 5.6 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
8CGF
 
 | | Interleukin-4 (wild type) pH 2.4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Loureiro-Ferreira, N, Mueller, T, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2023-02-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
3NH7
 
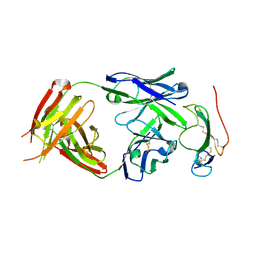 | | Crystal structure of the neutralizing Fab fragment AbD1556 bound to the BMP type I receptor IA | | Descriptor: | Antibody fragment Fab AbD1556, heavy chain, light chain, ... | | Authors: | Mueller, T.D, Harth, S, Sebald, W. | | Deposit date: | 2010-06-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A selection fit mechanism in BMP receptor IA as a possible source for BMP ligand-receptor promiscuity
Plos One, 5, 2010
|
|
5EPW
 
 | | C-Terminal Domain Of Human Coronavirus Nl63 Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Szelazek, B, Kabala, W, Kus, K, Zdzalik, M, Golik, P, Florek, D, Burmistrz, M, Pyrc, K, Dubin, G. | | Deposit date: | 2015-11-12 | | Release date: | 2017-02-22 | | Last modified: | 2017-05-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
2R53
 
 | |
2H64
 
 | |
2R52
 
 | |
8A4F
 
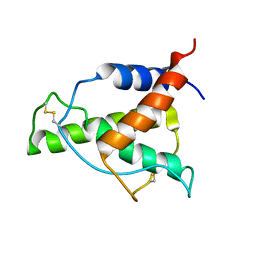 | | Human Interleukin-4 mutant - C3T-IL4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Mueller, T.D, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2022-06-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
3BMP
 
 | |
1REU
 
 | | Structure of the bone morphogenetic protein 2 mutant L51P | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, bone morphogenetic protein 2 | | Authors: | Keller, S, Nickel, J, Zhang, J.-L, Sebald, W, Mueller, T.D. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular recognition of BMP-2 and BMP receptor IA.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WAQ
 
 | |
1ES7
 
 | |
3QT2
 
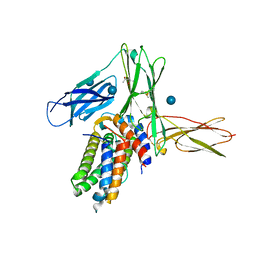 | | Structure of a cytokine ligand-receptor complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Interleukin-5, Interleukin-5 receptor subunit alpha, ... | | Authors: | Mueller, T.D, Patino, E, Kotzsch, A, Saremba, S, Nickel, J, Schmitz, W, Sebald, W. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure Analysis of the IL-5 Ligand-Receptor Complex Reveals a Wrench-like Architecture for IL-5Ralpha.
Structure, 19, 2011
|
|
1REW
 
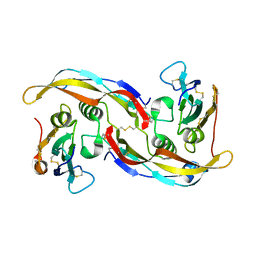 | | Structural refinement of the complex of bone morphogenetic protein 2 and its type IA receptor | | Descriptor: | bone morphogenetic protein 2, bone morphogenetic protein receptor type IA | | Authors: | Keller, S, Nickel, J, Zhang, J.-L, Sebald, W, Mueller, T.D. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Molecular recognition of BMP-2 and BMP receptor IA.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3QB4
 
 | |
3BK3
 
 | |
2CYK
 
 | |
2B8Z
 
 | | Crystal structure of the interleukin-4 variant R85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B90
 
 | | Crystal structure of the interleukin-4 variant T13DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8U
 
 | | Crystal structure of wildtype human Interleukin-4 | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8Y
 
 | | Crystal structure of the interleukin-4 variant T13DF82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
