2LSU
 
 | | The NMR high resolution structure of yeast Tah1 in a free form | | Descriptor: | TPR repeat-containing protein associated with Hsp90 | | Authors: | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
2LSV
 
 | | The NMR high resolution structure of yeast Tah1 in complex with the Hsp90 C-terminal tail | | Descriptor: | ATP-dependent molecular chaperone HSP82, TPR repeat-containing protein associated with Hsp90 | | Authors: | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | Deposit date: | 2012-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
5LM5
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM2 peptide (region 435-451) | | Descriptor: | DNA topoisomerase 2-associated protein PAT1, mRNA decapping protein 2 | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LMG
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM10 peptide (region 954-970) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LMF
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM3 peptide (region 484-500) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LON
 
 | | Structure of /K. lactis/ Dcp1-Dcp2 decapping complex. | | Descriptor: | KLLA0E01827p, KLLA0F23980p | | Authors: | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5LOP
 
 | | Structure of the active form of /K. lactis/ Dcp1-Dcp2-Edc3 decapping complex bound to m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, KLLA0A11308p, KLLA0E01827p, ... | | Authors: | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
2MJF
 
 | | Solution structure of the complex between the yeast Rsa1 and Hit1 proteins | | Descriptor: | Protein HIT1, Ribosome assembly 1 protein | | Authors: | Quinternet, M, Roth, B, Back, R, Jacquemin, C, Manival, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction.
Nucleic Acids Res., 42, 2014
|
|
6Y3Z
 
 | |
6Y3P
 
 | |
2MNJ
 
 | | NMR solution structure of the yeast Pih1 and Tah1 C-terminal domains complex | | Descriptor: | Protein interacting with Hsp90 1, TPR repeat-containing protein associated with Hsp90 | | Authors: | Quinternet, M, Jacquemin, C, Charpentier, B, Manival, X. | | Deposit date: | 2014-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure/Function Analysis of Protein-Protein Interactions Developed by the Yeast Pih1 Platform Protein and Its Partners in Box C/D snoRNP Assembly.
J.Mol.Biol., 427, 2015
|
|
2M3F
 
 | | NMR structure of Rsa1p238-259 from S. Cerevisiae | | Descriptor: | Ribosome assembly 1 protein | | Authors: | Quinternet, M, Manival, X. | | Deposit date: | 2013-01-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly.
Nucleic Acids Res., 42, 2014
|
|
8C7M
 
 | |
8CR6
 
 | | mouse Interleukin-12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 subunit alpha, Interleukin-12 subunit beta, ... | | Authors: | Merceron, R, Felix, J, Lambert, E, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8CR5
 
 | | Murine Interleukin-12 receptor beta 1 domain 1 in complex with murine Interleukin-12 beta. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, Interleukin-12 subunit beta, ... | | Authors: | Merceron, R, Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8CR8
 
 | | human Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, TERBIUM(III) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ODZ
 
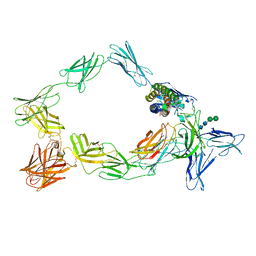 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OE4
 
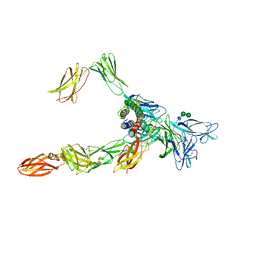 | | Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8ODX
 
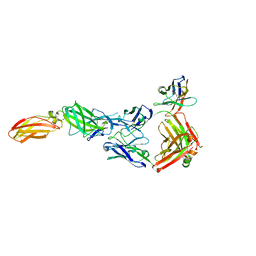 | |
8OE0
 
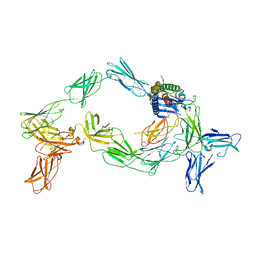 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 2). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PB1
 
 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1), obtained after local refinement. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-06-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8COO
 
 | | Solution structure of Zipcode binding protein 1 (ZBP1) KH3(DD)KH4 domains in complex with N6-Methyladenosine containing RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 1, RNA_(5'-R(*(UP*CP*GP*GP*(6MZ)P*CP*U)-3') | | Authors: | Nicastro, G, Abis, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-07 | | Method: | SOLUTION NMR | | Cite: | Direct m6A recognition by IMP1 underlays an alternative model of target selection for non-canonical methyl-readers.
Nucleic Acids Res., 51, 2023
|
|
8Q5B
 
 | |
