5LUE
 
 | | Minor form of the recombinant cytotoxin-1 from N. oxiana | | Descriptor: | VC-1=CYTOTOXIN | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
7B3J
 
 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
7B3K
 
 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
5LV6
 
 | | N-terminal motif dimerization of EGFR transmembrane domain in bicellar environment | | Descriptor: | Epidermal growth factor receptor | | Authors: | Bragin, P, Bocharov, E, Mineev, K, Bocharova, O, Arseniev, A. | | Deposit date: | 2016-09-12 | | Release date: | 2017-04-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Conformation of the Epidermal Growth Factor Receptor Transmembrane Domain Dimer Dynamically Adapts to the Local Membrane Environment.
Biochemistry, 56, 2017
|
|
8BVC
 
 | |
5NAM
 
 | |
5NAO
 
 | |
2MQU
 
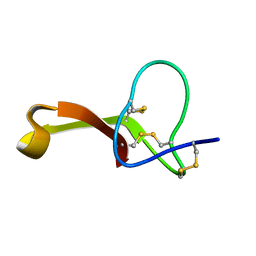 | | Spatial structure of Hm-3, a membrane-active spider toxin affecting sodium channels | | Descriptor: | Neurotoxin Hm-3 | | Authors: | Myshkin, M.Y, Shenkarev, Z.O, Paramonov, A.S, Berkut, A.A, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2014-06-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of membrane-active toxin from crab spider Heriaeus melloteei suggests parallel evolution of sodium channel gating modifiers in Araneomorphae and Mygalomorphae.
J. Biol. Chem., 290, 2015
|
|
5T8A
 
 | | Recombinant cytotoxin-I from the venom of cobra N. oxiana | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
2M1F
 
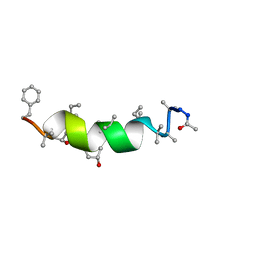 | | NMR Structure of Antiamoebin I (peptaibol antibiotic) bound to DMPC/DHPC bicelles | | Descriptor: | Antiamoebin I | | Authors: | Shenkarev, Z.O, Paramonov, A.S, Gizatullina, A.K. | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2013-10-02 | | Method: | SOLUTION NMR | | Cite: | Peptaibol antiamoebin I: spatial structure, backbone dynamics, interaction with bicelles and lipid-protein nanodiscs, and pore formation in context of barrel-stave model.
Chem.Biodivers., 10, 2013
|
|
2LOH
 
 | |
2MJO
 
 | |
2LZO
 
 | | Spatial structure of Pi-AnmTX Ugr 9a-1 | | Descriptor: | UGTX | | Authors: | Mineev, K, Arseniev, A. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Sea Anemone Peptide with Uncommon beta-Hairpin Structure Inhibits Acid-sensing Ion Channel 3 (ASIC3) and Reveals Analgesic Activity.
J.Biol.Chem., 288, 2013
|
|
2MET
 
 | | NMR spatial structure of the trimeric mutant TM domain of VEGFR2 receptor. | | Descriptor: | Vascular endothelial growth factor receptor 2 | | Authors: | Mineev, K.S, Arseniev, A.A, Shulepko, M, Lyukmanova, E.N, Kirpichnikov, M.P. | | Deposit date: | 2013-10-02 | | Release date: | 2014-07-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of alternative transmembrane domain conformations in VEGF receptor 2 activation
Structure, 22, 2014
|
|
2N2A
 
 | | Spatial structure of HER2/ErbB2 dimeric transmembrane domain in the presence of cytoplasmic juxtamembrane domains | | Descriptor: | Receptor tyrosine-protein kinase erbB-2 | | Authors: | Bragin, P.E, Mineev, K.S, Bocharov, E, Bocharova, O, Arseniev, A. | | Deposit date: | 2015-05-05 | | Release date: | 2016-02-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | HER2 Transmembrane Domain Dimerization Coupled with Self-Association of Membrane-Embedded Cytoplasmic Juxtamembrane Regions.
J.Mol.Biol., 428, 2016
|
|
2N5S
 
 | |
2MZF
 
 | | Purotoxin-2 NMR structure in water | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Vassilevski, A, Oparin, P, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2N90
 
 | |
2N85
 
 | | NMR structure of OtTx1a - AMP in DPC micelles | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
2N86
 
 | | NMR structure of OtTx1a - ICK | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
2MZG
 
 | | Purotoxin-2 NMR structure in DPC micelles | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Oparin, P, Vassilevski, A, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2MIC
 
 | |
6RC7
 
 | | NMR structure of cytotoxin 3 from Naja kaouthia in solution, major form | | Descriptor: | Cytotoxin 3 | | Authors: | Dubinnyi, M.A, Dubovskii, P.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2019-04-11 | | Release date: | 2020-05-13 | | Last modified: | 2021-05-26 | | Method: | SOLUTION NMR | | Cite: | The omega-loop of cobra cytotoxins tolerates multiple amino acid substitutions.
Biochem.Biophys.Res.Commun., 558, 2021
|
|
7AY8
 
 | | NMR solution structure of Tbo-IT2 | | Descriptor: | Tbo-IT2 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Korolkova, Y.A, Maleeva, E.E, Andreev, Y.A, Kozlov, S.A, Arseniev, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold.
Toxins, 13, 2021
|
|
1G2G
 
 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
