8V4Y
 
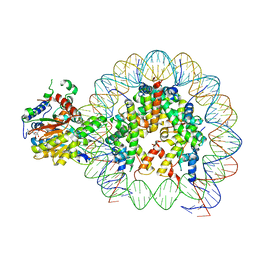 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Histone H2A type 1, ... | | 著者 | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | 登録日 | 2023-11-29 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V7L
 
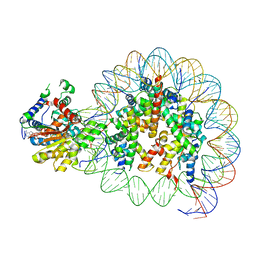 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | 著者 | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | 登録日 | 2023-12-04 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V6V
 
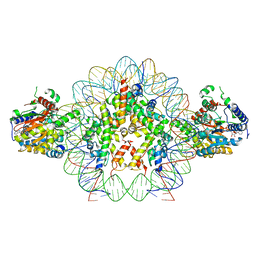 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | 著者 | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | 登録日 | 2023-12-03 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8EIR
 
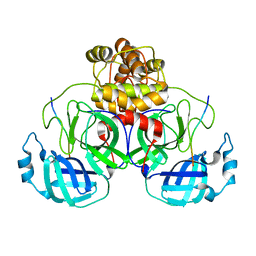 | | SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction | | 分子名称: | 3C-like proteinase nsp5, nsp7-nsp10 of Replicase polyprotein 1a | | 著者 | Narwal, M, Edwards, T, Armache, J.P, Murakami, K.S. | | 登録日 | 2022-09-15 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | SARS-CoV-2 polyprotein substrate regulates the stepwise M pro cleavage reaction.
J.Biol.Chem., 299, 2023
|
|
8EKE
 
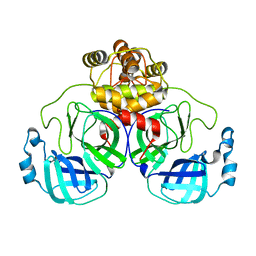 | |
2WWA
 
 | | Cryo-EM structure of idle yeast Ssh1 complex bound to the yeast 80S ribosome | | 分子名称: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | 著者 | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2018-10-03 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
2WWB
 
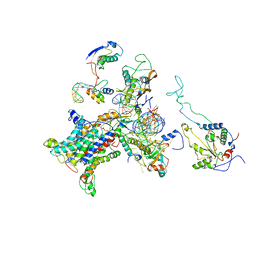 | | CRYO-EM STRUCTURE OF THE MAMMALIAN SEC61 COMPLEX BOUND TO THE ACTIVELY TRANSLATING WHEAT GERM 80S RIBOSOME | | 分子名称: | 25S RRNA, 5.8S RRNA, 60S RIBOSOMAL PROTEIN L17-A, ... | | 著者 | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2018-10-03 | | 実験手法 | ELECTRON MICROSCOPY (6.48 Å) | | 主引用文献 | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
2WW9
 
 | | Cryo-EM structure of the active yeast Ssh1 complex bound to the yeast 80S ribosome | | 分子名称: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | 著者 | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | 登録日 | 2009-10-22 | | 公開日 | 2009-12-08 | | 最終更新日 | 2018-10-03 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | 著者 | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2010-10-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6NE3
 
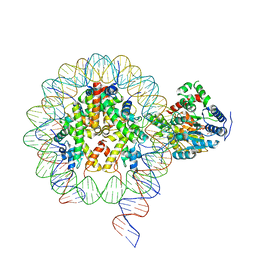 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h bound at SHL-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (156-MER), Histone H2A type 1, ... | | 著者 | Armache, J.-P, Gamarra, N, Johnson, S.L, Leonard, J.D, Wu, S, Narlikar, G.N, Cheng, Y. | | 登録日 | 2018-12-16 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome.
Elife, 8, 2019
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | 分子名称: | rRNA expansion segment ES27L in an "out" conformation | | 著者 | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2010-10-13 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V5H
 
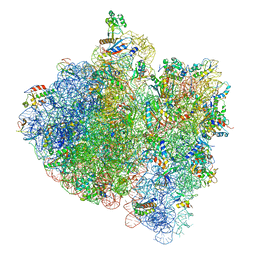 | | E.Coli 70s Ribosome Stalled During Translation Of Tnac Leader Peptide. | | 分子名称: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Seidelt, B, Innis, C.A, Wilson, D.N, Gartmann, M, Armache, J, Villa, E, Trabuco, L.G, Becker, T, Mielke, T, Schulten, K, Steitz, T.A, Beckmann, R. | | 登録日 | 2009-10-26 | | 公開日 | 2014-07-09 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Structural insight into nascent polypeptide chain-mediated translational stalling.
Science, 326, 2009
|
|
7K6Q
 
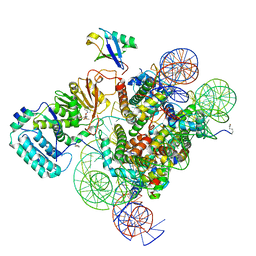 | | Active state Dot1 bound to the H4K16ac nucleosome | | 分子名称: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | 登録日 | 2020-09-21 | | 公開日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
7K6P
 
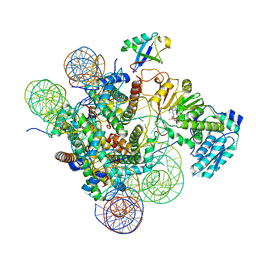 | | Active state Dot1 bound to the unacetylated H4 nucleosome | | 分子名称: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Truong, D.M, Lee, R, Armache, J.-P, Boeke, J.D, Armache, K.-J. | | 登録日 | 2020-09-21 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation.
Science, 371, 2021
|
|
8G57
 
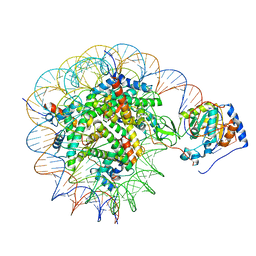 | | Structure of nucleosome-bound Sirtuin 6 deacetylase | | 分子名称: | DNA strand 1, DNA strand 2, Histone H2A type 1-B/E, ... | | 著者 | Chio, U.S, Rechiche, O, Bryll, A.R, Zhu, J, Feldman, J.L, Peterson, C.L, Tan, S, Armache, J.-P. | | 登録日 | 2023-02-11 | | 公開日 | 2023-04-26 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Cryo-EM structure of the human Sirtuin 6-nucleosome complex.
Sci Adv, 9, 2023
|
|
6CX0
 
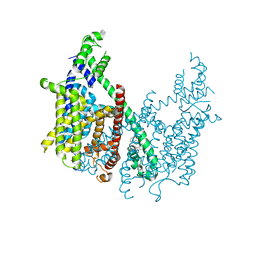 | | Structure of AtTPC1 D376A | | 分子名称: | (1S,3R)-1-(3-{[4-(2-fluorophenyl)piperazin-1-yl]methyl}-4-methoxyphenyl)-2,3,4,9-tetrahydro-1H-beta-carboline-3-carboxylic acid, CALCIUM ION, Two pore calcium channel protein 1 | | 著者 | Kintzer, A.F, Stroud, R.M. | | 登録日 | 2018-04-02 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7TN2
 
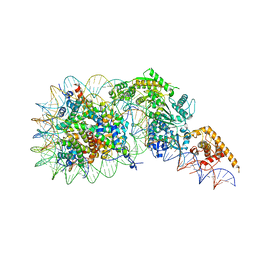 | | Composite model of a Chd1-nucleosome complex in the nucleotide-free state derived from 2.3A and 2.7A Cryo-EM maps | | 分子名称: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | 著者 | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | 登録日 | 2022-01-20 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
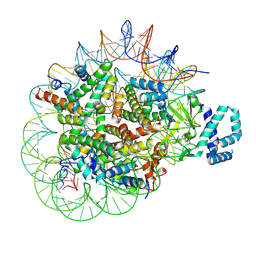
8T9F
 
 | |
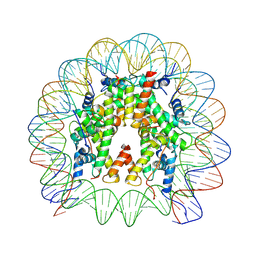
8THU
 
 | |
7SWY
 
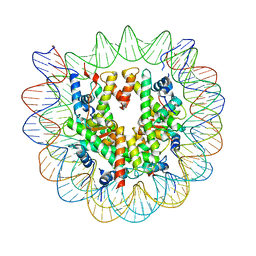 | | 2.6 A structure of a 40-601[TA-rich+1]-40 nucleosome | | 分子名称: | DNA Guide Strand, DNA Tracking Strand, Histone H2A, ... | | 著者 | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | 登録日 | 2021-11-21 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6SCT
 
 | | Cryo-EM structure of the consensus triskelion hub of the clathrin coat complex | | 分子名称: | Clathrin heavy chain, Clathrin light chain | | 著者 | Morris, K.L, Cameron, A.D, Sessions, R, Smith, C.J. | | 登録日 | 2019-07-25 | | 公開日 | 2019-10-02 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.69 Å) | | 主引用文献 | Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3IZQ
 
 | | Structure of the Dom34-Hbs1-GDPNP complex bound to a translating ribosome | | 分子名称: | Elongation factor 1 alpha-like protein, Protein DOM34 | | 著者 | Becker, T, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Sieber, H, Abdel Motaal, B, Mielke, T, Berninghausen, O, Beckmann, R. | | 登録日 | 2010-11-30 | | 公開日 | 2011-06-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | 著者 | O'Neill, A.G, Kollman, J.M. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G8F
 
 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | 著者 | O'Neill, A.G, Kollman, J.M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
6E1M
 
 | | Structure of AtTPC1(DDE) reconstituted in saposin A | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | 著者 | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | 登録日 | 2018-07-10 | | 公開日 | 2018-09-19 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
