3A3Z
 
 | | Crystal structure of the human VDR ligand binding domain bound to the synthetic agonist compound 2alpha-methyl-AMCR277A(C23S) | | Descriptor: | (1S,2S,3R,5Z,7E,14beta,17alpha)-17-[(2S,4S)-4-(2-hydroxy-2-methylpropyl)-2-methyltetrahydrofuran-2-yl]-2-methyl-9,10-secoandrosta-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Antony, P, Huet, T, Sigueiro, R, Rochel, N, Moras, D, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2009-06-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3
J.Med.Chem., 53, 2010
|
|
3A40
 
 | | Crystal structure of the human VDR ligand binding domain bound to the synthetic agonist compound 2alpha-methyl-AMCR277B(C23R) | | Descriptor: | (1S,2S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-2-methyl-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Antony, P, Huet, T, Sigueiro, R, Rochel, N, Moras, D, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2009-06-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3
J.Med.Chem., 53, 2010
|
|
3CS6
 
 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | Authors: | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, P, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
3CS4
 
 | | Structure-based design of a superagonist ligand for the vitamin D nuclear receptor | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha)-17-[(2S,4S)-4-(2-hydroxy-2-methylpropyl)-2-methyltetrahydrofuran-2-yl]-9,10-secoandrosta-5,7,10-triene-1,3-diol, Vitamin D3 receptor | | Authors: | Hourai, S, Rodriguez, L.C, Antony, P, Reina-San-Martin, B, Ciesielski, F, Magnier, B.C, Schoonjans, K, Mourino, A, Rochel, N, Moras, D. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a superagonist ligand for the vitamin d nuclear receptor.
Chem.Biol., 15, 2008
|
|
2ZY0
 
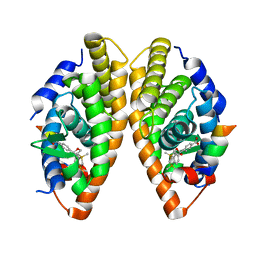 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-1,3-benzodisilol-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
2ZXZ
 
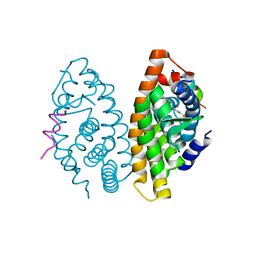 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-inden-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
3A78
 
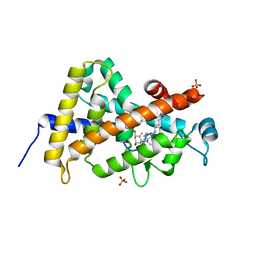 | | Crystal structure of the human VDR ligand binding domain bound to the natural metabolite 1alpha,25-dihydroxy-3-epi-vitamin D3 | | Descriptor: | (1S,3S,5Z,7E,14beta,17alpha)-9,10-secocholesta-5,7,10-triene-1,3,25-triol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Sigueiro, R, Antony, P, Rochel, N, Moras, D. | | Deposit date: | 2009-09-18 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the activation of the human Vitamin D receptor by the natural metabolite 1alpha,25-dihydroxy-3-epi-vitamin D3
To be Published
|
|
4RUJ
 
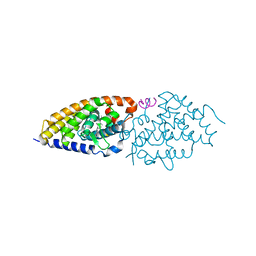 | | Crystal structure of zVDR L337H mutant-VD complex | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
4RUO
 
 | | Crystal structure of zVDR L337H mutant-gemini complex | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
4RUP
 
 | | Crystal structure of zVDR L337H mutant-Gemini72 complex | | Descriptor: | (1R,3R,7E,17beta)-17-[(1R)-6,6,6-trifluoro-5-hydroxy-1-(4-hydroxy-4-methylpentyl)-5-(trifluoromethyl)hex-3-yn-1-yl]-9,1 0-secoestra-5,7-diene-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
3A9E
 
 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | Authors: | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-10-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
8ROL
 
 | |
8RO9
 
 | | Human cohesin SMC1A-HD(longCC-EQ)/RAD21-C complex - Open/closed P-loop conformation | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 64-kDa C-terminal product, Structural maintenance of chromosomes protein 1A | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROE
 
 | |
8ROA
 
 | |
8ROK
 
 | |
8ROH
 
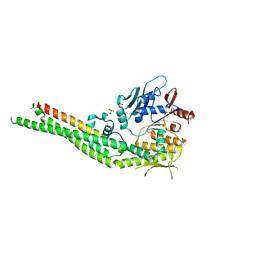 | | Human cohesin SMC3-HD(EQ)/RAD21-C complex - Apo conformation | | Descriptor: | Double-strand-break repair protein rad21 homolog, GLYCEROL, SULFATE ION, ... | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROF
 
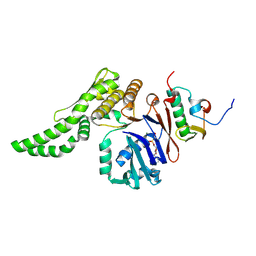 | |
8ROC
 
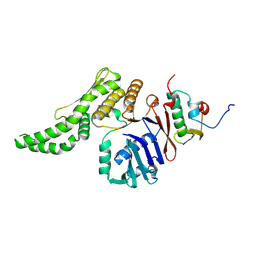 | |
8ROJ
 
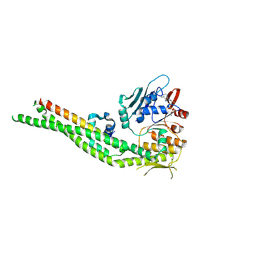 | | Human cohesin SMC3-HD/RAD21-C complex - ADP-Mg-bound conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Vitoria Gomes, M, Romier, C. | | Deposit date: | 2024-01-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The cohesin ATPase cycle is mediated by specific conformational dynamics and interface plasticity of SMC1A and SMC3 ATPase domains.
Cell Rep, 43, 2024
|
|
8ROB
 
 | |
8RO6
 
 | |
8RO8
 
 | |
8ROD
 
 | |
8ROG
 
 | |
