1N3I
 
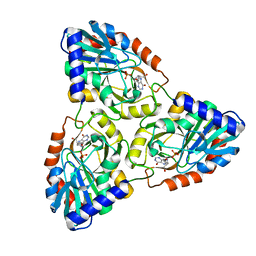 | | Crystal Structure of Mycobacterium tuberculosis PNP with transition state analog DADMe-ImmH | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine Nucleoside Phosphorylase | | Authors: | Lewandowicz, A, Shi, W, Evans, G.B, Tyler, P.C, Furneaux, R.H, Basso, L.A, Santos, D.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-10-28 | | Release date: | 2003-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Over-The-Barrier Transition State Analogues Provide New Chemistries for Inhibitor Design: The Case of Purine Nucleoside Phosphorylase
BIOCHEMISTRY, 42, 2003
|
|
5EOC
 
 | | Crystal structure of Fab C2 in complex with a Cyclic variant of Hepatitis C Virus E2 epitope I | | Descriptor: | ALA-CYS-GLN-LEU-ILE-ASN-THR-ASN-GLY-SER-TRP-HIS-ILE-CYS, Fab fragment (Heavy chain), Fab fragment (Light chain) | | Authors: | Berisio, R, Ruggiero, A, Sandomenico, A, Patel, A.H, Ruvo, M, Vitagliano, L. | | Deposit date: | 2015-11-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generation and Characterization of Monoclonal Antibodies against a Cyclic Variant of Hepatitis C Virus E2 Epitope 412-422.
J.Virol., 90, 2016
|
|
1QGI
 
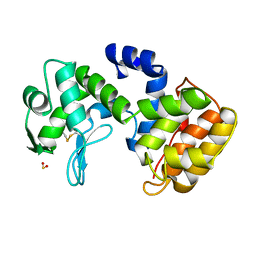 | | CHITOSANASE FROM BACILLUS CIRCULANS | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CHITOSANASE), SULFATE ION | | Authors: | Saito, J, Kita, A, Higuchi, Y, Nagata, Y, Ando, A, Miki, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism.
J.Biol.Chem., 274, 1999
|
|
2D05
 
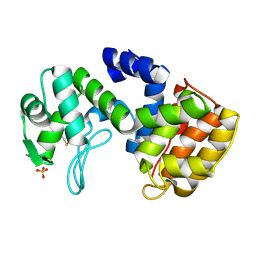 | | Chitosanase From Bacillus circulans mutant K218P | | Descriptor: | Chitosanase, SULFATE ION | | Authors: | Fukamizo, T, Amano, S, Yamaguchi, K, Yoshikawa, T, Katsumi, T, Saito, J, Suzuki, M, Miki, K, Nagata, Y, Ando, A. | | Deposit date: | 2005-07-25 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacillus circulans MH-K1 Chitosanase: Amino Acid Residues Responsible for Substrate Binding
J.Biochem.(Tokyo), 138, 2005
|
|
5HWA
 
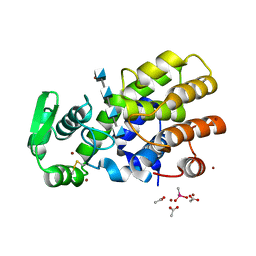 | | Crystal Structure of MH-K1 chitosanase in substrate-bound form | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Suzuki, M, Saito, A, Ando, A, Miki, K, Saito, J. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1 in complex with chitotetraose
Biomed.Biochim.Acta, 1868, 2024
|
|
8YTD
 
 | | Crystal Structure of TrkA D5 domain in complex with two different macrocyclic peptides | | Descriptor: | 1,2-ETHANEDIOL, High affinity nerve growth factor receptor, Macrocyclic Peptide | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
8YTE
 
 | | Crystal Structure of TrkA D5 domain in complex with macrocyclic peptide | | Descriptor: | 1,2-ETHANEDIOL, AMINOMETHYLAMIDE, High affinity nerve growth factor receptor, ... | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
5EH0
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, N2-(2-Methoxy-4-(1-methyl-1H-pyrazol-4-yl)phenyl)-N8-neopentylpyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-27 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
5EI2
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-(2,4-dimethoxyphenyl)-8-(1-methylpyrazol-4-yl)pyrido[3,4-d]pyrimidin-2-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
5EHL
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Dual specificity protein kinase TTK | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach
To Be Published
|
|
5EHY
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 4-(furan-3-yl)-3-phenyl-2~{H}-pyrazolo[4,3-c]pyridine, ... | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
5EI8
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}-[2-methoxy-4-(1-methylpyrazol-4-yl)phenyl]-8-(1-methylpyrazol-4-yl)pyrido[3,4-d]pyrimidin-2-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
5EHO
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}8-cyclohexyl-~{N}2-[2-methoxy-4-(1-methylpyrazol-4-yl)phenyl]pyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach
To Be Published
|
|
4C4E
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, N-(3,4-dimethoxyphenyl)-2-(1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridin-6-amine, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C4F
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
5EI6
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}-(2,4-dimethoxyphenyl)-5-(1-methylpyrazol-4-yl)isoquinolin-3-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
4C4G
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C4J
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(1-methyl-1H-imidazol-5-yl)phenyl]amino}-2-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C4H
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-((2-chloro-4-(dimethylcarbamoyl)phenyl)amino)-2-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
4C4I
 
 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, tert-butyl 6-{[2-chloro-4-(dimethylcarbamoyl)phenyl]amino}-2-(1,3-oxazol-5-yl)-1H-pyrrolo[3,2-c]pyridine-1-carboxylate | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
1PHS
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE SEED STORAGE PROTEIN PHASEOLIN AT 3 ANGSTROMS RESOLUTION | | Descriptor: | PHASEOLIN, BETA-TYPE PRECURSOR | | Authors: | Lawrence, M.C, Suzuki, E, Varghese, J.N, Davis, P.C, Vandonkelaar, A, Tulloch, P.A, Colman, P.M. | | Deposit date: | 1990-03-21 | | Release date: | 1990-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structure of the seed storage protein phaseolin at 3 A resolution.
EMBO J., 9, 1990
|
|
2ASK
 
 | | Structure of human Artemin | | Descriptor: | SULFATE ION, artemin | | Authors: | Silvian, L, Jin, P, Carmillo, P, Boriack-Sjodin, P.A, Pelletier, C, Rushe, M, Gong, B.J, Sah, D, Pepinsky, B, Rossomando, A. | | Deposit date: | 2005-08-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Artemin crystal structure reveals insights into heparan sulfate binding.
Biochemistry, 45, 2006
|
|
1GND
 
 | | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR, ALPHA-ISOFORM | | Descriptor: | GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Authors: | Schalk, I, Zeng, K, Wu, S.-K, Stura, E.A, Metteson, J, Huang, M, Tandon, A, Wilson, I.A, Balch, W.E. | | Deposit date: | 1996-07-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure and mutational analysis of Rab GDP-dissociation inhibitor.
Nature, 381, 1996
|
|
1NW4
 
 | | Crystal Structure of Plasmodium falciparum Purine Nucleoside Phosphorylase in complex with ImmH and Sulfate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Shi, W, Ting, L.M, Kicska, G.A, Lewandowicz, A, Tyler, P.C, Evans, G.B, Furneaux, R.H, Kim, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-02-05 | | Release date: | 2004-03-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plasmodium falciparum Purine Nucleoside Phosphorylase: CRYSTAL STRUCTURES, IMMUCILLIN INHIBITORS, AND DUAL CATALYTIC FUNCTION.
J.Biol.Chem., 279, 2004
|
|
1Q1G
 
 | | Crystal structure of Plasmodium falciparum PNP with 5'-methylthio-immucillin-H | | Descriptor: | 3,4-DIHYDROXY-2-[(METHYLSULFANYL)METHYL]-5-(4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)PYRROLIDINIUM, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Shi, W, Ting, L.M, Kicska, G.A, Lewandowicz, A, Tyler, P.C, Evans, G.B, Furneaux, R.H, Kim, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-07-19 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Plasmodium falciparum Purine Nucleoside Phosphorylase: CRYSTAL STRUCTURES, IMMUCILLIN INHIBITORS, AND DUAL CATALYTIC FUNCTION.
J.Biol.Chem., 279, 2004
|
|
