7X5D
 
 | |
5K2J
 
 | |
5K2I
 
 | |
5K1G
 
 | | Crystal structure of reduced Prx3 from Vibrio vulnificus | | Descriptor: | 1-Cys peroxiredoxin | | Authors: | Ahn, J, Ha, N.-C. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal structure of peroxiredoxin 3 fromVibrio vulnificusand its implications for scavenging peroxides and nitric oxide.
IUCrJ, 5, 2018
|
|
7YVD
 
 | |
7DTG
 
 | |
7CRG
 
 | |
6JLB
 
 | |
7D9N
 
 | |
1M22
 
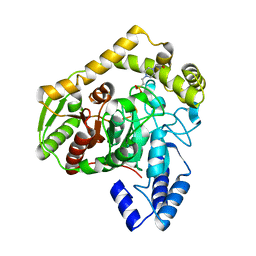 | | X-ray structure of native peptide amidase from Stenotrophomonas maltophilia at 1.4 A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, peptide amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1M21
 
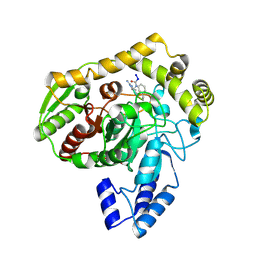 | | Crystal structure analysis of the peptide amidase PAM in complex with the competitive inhibitor chymostatin | | Descriptor: | CHYMOSTATIN, Peptide Amidase | | Authors: | Labahn, J, Neumann, S, Buldt, G, Kula, M.-R, Granzin, J. | | Deposit date: | 2002-06-21 | | Release date: | 2002-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An alternative mechanism for amidase signature enzymes
J.MOL.BIOL., 322, 2002
|
|
1MPG
 
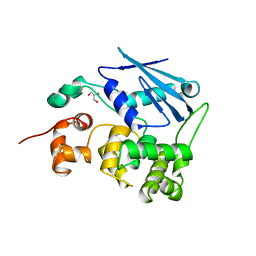 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
6MIU
 
 | |
6MJ7
 
 | | Crystal structure of p62 ZZ domain in complex with free arginine | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ARGININE, Sequestosome-1, ... | | Authors: | Ahn, J, Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2018-09-20 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | ZZ-dependent regulation of p62/SQSTM1 in autophagy.
Nat Commun, 9, 2018
|
|
6O5W
 
 | | Crystal structure of MORC3 CW domain fused with viral influenza A NS1 peptide | | Descriptor: | NS1-linked peptide,MORC family CW-type zinc finger protein 3, ZINC ION | | Authors: | Ahn, J, Zhang, Y, Vann, K.R, Kutateleadze, T. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | MORC3 Is a Target of the Influenza A Viral Protein NS1.
Structure, 27, 2019
|
|
4QCJ
 
 | | Crystal Structure of OdhI from Corynebacterium glutamicum | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Labahn, J, Raasch, K, Eggeling, L, Bocola, M, Leitner, A, Bott, M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction of 2-oxoglutarate dehydrogenase OdhA with its inhibitor OdhI in Corynebacterium glutamicum: Mutants and a model.
J.Biotechnol., 191, 2014
|
|
5JK7
 
 | | The X-ray structure of the DDB1-DCAF1-Vpr-UNG2 complex | | Descriptor: | DNA damage-binding protein 1, Protein VPRBP, Protein Vpr, ... | | Authors: | Calero, G, Ahn, J, Wu, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
4BZB
 
 | | Crystal structure of the tetrameric dGTP-bound SAMHD1 mutant catalytic core | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BZC
 
 | | Crystal structure of the tetrameric dGTP-bound wild type SAMHD1 catalytic core | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION, ... | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
7TA8
 
 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
4Z8L
 
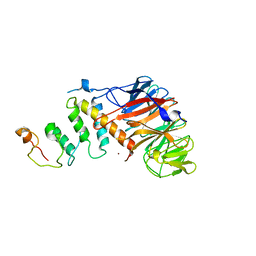 | | Crystal structure of DCAF1/SIV-MND VPX/MND SAMHD1 NTD ternary complex | | Descriptor: | Protein VPRBP, SAM domain and HD domain-containing protein, Vpx protein, ... | | Authors: | Koharudin, L.M, Wu, Y, Calero, G, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2015-04-09 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile alpha Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors.
J.Biol.Chem., 290, 2015
|
|
2KOD
 
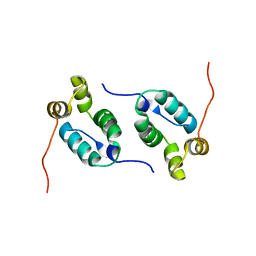 | | A high-resolution NMR structure of the dimeric C-terminal domain of HIV-1 CA | | Descriptor: | HIV-1 CA C-terminal domain | | Authors: | Byeon, I.-J.L, Jung, J, Ahn, J, concel, J, Gronenborn, A.M. | | Deposit date: | 2009-09-18 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function.
Cell(Cambridge,Mass.), 139, 2009
|
|
2M65
 
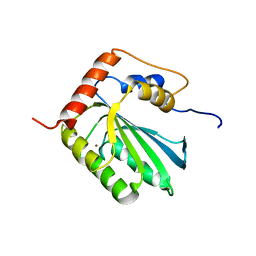 | | NMR structure of human restriction factor APOBEC3A | | Descriptor: | Probable DNA dC->dU-editing enzyme APOBEC-3A, ZINC ION | | Authors: | Byeon, I.L, Byeon, C, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity.
Nat Commun, 4, 2013
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
