6D0M
 
 | | Polymerase Eta post-insertion binary complex with cytarabine (AraC) | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*CP*AP*GP*TP*GP*CP*T)-3'), DNA polymerase eta, DNA/RNA (5'-D(*AP*GP*CP*AP*CP*TP*GP*T)-R(P*(CAR))-3'), ... | | Authors: | Rechkoblit, O, Aggarwal, A.K. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Structural basis for polymerase eta-promoted resistance to the anticancer nucleoside analog cytarabine.
Sci Rep, 8, 2018
|
|
6D0Z
 
 | | Polymerase Eta cytarabine (AraC) extension ternary complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*TP*TP*GP*CP*AP*GP*TP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Rechkoblit, O, Aggarwal, A.K. | | Deposit date: | 2018-04-11 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for polymerase eta-promoted resistance to the anticancer nucleoside analog cytarabine.
Sci Rep, 8, 2018
|
|
7S0T
 
 | | Structure of DNA polymerase zeta with mismatched DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Malik, R, Ubarretxena, I.B, Aggarwal, A.K. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structure of translesion DNA synthesis polymerase zeta with a base pair mismatch.
Nat Commun, 13, 2022
|
|
7TW9
 
 | |
3BJY
 
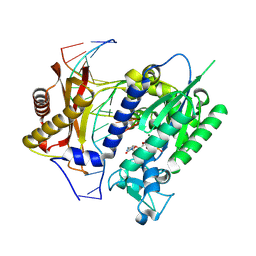 | | Catalytic core of Rev1 in complex with DNA (modified template guanine) and incoming nucleotide | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*DAP*DTP*DCP*DCP*DTP*DCP*DCP*DCP*DCP*DTP*DAP*(DOC))-3'), DNA (5'-D(*DTP*DAP*DAP*(P)P*DGP*DTP*DAP*DGP*DGP*DGP*DGP*DAP*DGP*DGP*DAP*DT)-3'), ... | | Authors: | Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2007-12-05 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase
Structure, 16, 2008
|
|
3BSB
 
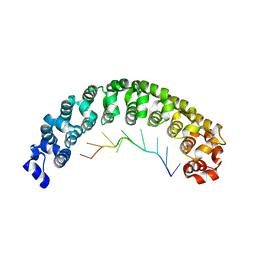 | | Crystal Structure of Human Pumilio1 in Complex with CyclinB reverse RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*AP*UP*GP*UP*U)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-23 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
2BAM
 
 | |
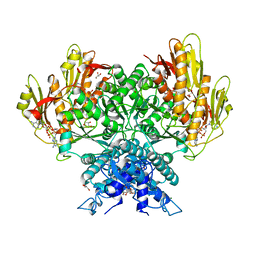
8FMF
 
 | |
8FM1
 
 | |
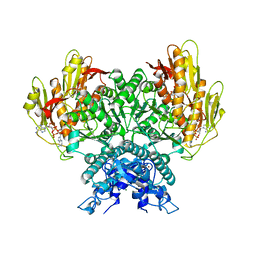
8FMG
 
 | |
8FMH
 
 | |
7TW7
 
 | |
7TW8
 
 | |
1OXJ
 
 | |
1N72
 
 | | Structure and Ligand of a Histone Acetyltransferase Bromodomain | | Descriptor: | HISTONE ACETYLTRANSFERASE | | Authors: | Dhalluin, C, Carlson, J.E, Zeng, L, He, C, Aggarwal, A.K, Zhou, M.-M. | | Deposit date: | 2002-11-12 | | Release date: | 2002-12-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure and Ligand of a Histone Acetyltransferase Bromodomain
Nature, 399, 1999
|
|
1MX3
 
 | | Crystal structure of CtBP dehydrogenase core holo form | | Descriptor: | ACETIC ACID, C-terminal binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, V, Carlson, J.E, Ohgi, K.E, Edwards, T.E, Rose, D.W, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2002-10-01 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transcription Corepressor CtBP Is an NAD+-Regulated Dehydrogenase
Mol.Cell, 10, 2002
|
|
1JIH
 
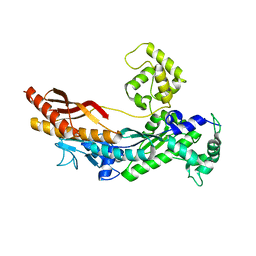 | | Yeast DNA Polymerase ETA | | Descriptor: | DNA Polymerase ETA | | Authors: | Trincao, J, Johnson, R.E, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2001-07-02 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis
Mol.Cell, 8, 2001
|
|
1JGG
 
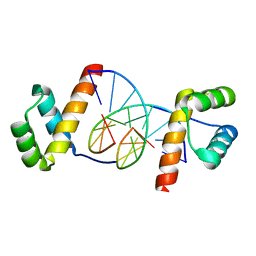 | | Even-skipped Homeodomain Complexed to AT-rich DNA | | Descriptor: | 5'-D(P*AP*AP*TP*TP*CP*AP*AP*TP*TP*A)-3', 5'-D(P*TP*AP*AP*TP*TP*GP*AP*AP*TP*T)-3', Segmentation Protein Even-Skipped | | Authors: | Hirsch, J.A, Aggarwal, A.K. | | Deposit date: | 2001-06-25 | | Release date: | 2001-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity.
EMBO J., 14, 1995
|
|
1DFM
 
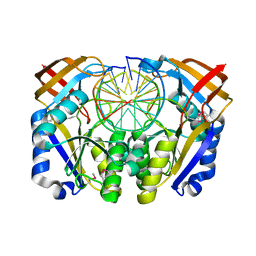 | | Crystal structure of restriction endonuclease BGLII complexed with DNA 16-mer | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), ENDONUCLEASE BGLII | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-21 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
1ES8
 
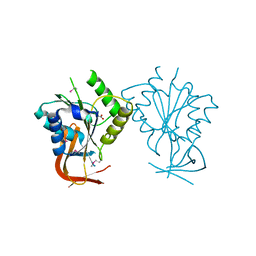 | | Crystal structure of free BglII | | Descriptor: | ACETIC ACID, RESTRICTION ENDONUCLEASE BGLII | | Authors: | Lukacs, C.M, Aggarwal, A.K. | | Deposit date: | 2000-04-07 | | Release date: | 2001-01-17 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of free BglII reveals an unprecedented scissor-like motion for opening an endonuclease.
Nat.Struct.Biol., 8, 2001
|
|
1D2I
 
 | | CRYSTAL STRUCTURE OF RESTRICTION ENDONUCLEASE BGLII COMPLEXED WITH DNA 16-MER | | Descriptor: | DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), MAGNESIUM ION, PROTEIN (RESTRICTION ENDONUCLEASE BGLII) | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
1Q7F
 
 | |
1S97
 
 | | DPO4 with GT mismatch | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*G)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SDO
 
 | | Crystal Structure of Restriction Endonuclease BstYI | | Descriptor: | BstYI | | Authors: | Townson, S.A, Samuelson, J.C, Vanamee, E.S, Edwards, T.A, Escalante, C.R, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2004-02-13 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of BstYI at 1.85 A Resolution: A Thermophilic Restriction Endonuclease with Overlapping Specificities to BamHI and BglII
J.Mol.Biol., 338, 2004
|
|
1S9F
 
 | | DPO with AT matched | | Descriptor: | 2',3'-DIDEOXYCYTOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
