7QP6
 
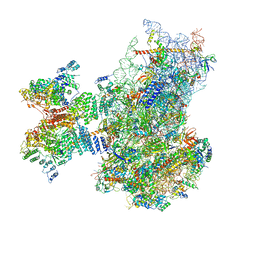 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | Deposit date: | 2022-01-03 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
2MH5
 
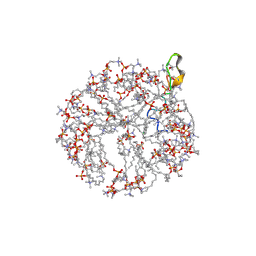 | | Structure and NMR assignments of lantibiotic NAI-107 in DPC micelles | | Descriptor: | Lantibiotic 107891, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Munch, D, Muller, A, Schneider, T, Kohl, B, Wenzel, M, Bandow, J, Maffioli, S, Sosio, M, Donadio, S, Wimmer, R, Sahl, H. | | Deposit date: | 2013-11-18 | | Release date: | 2014-03-05 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The Lantibiotic NAI-107 Binds to Bactoprenol-bound Cell Wall Precursors and Impairs Membrane Functions.
J.Biol.Chem., 289, 2014
|
|
6FBV
 
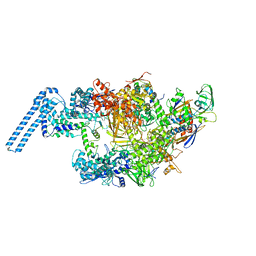 | | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Das, K, Lin, W, Ebright, E. | | Deposit date: | 2017-12-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
6ASG
 
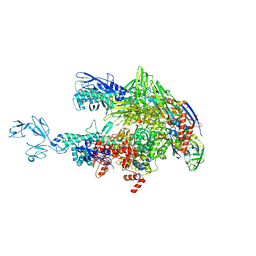 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
5UAO
 
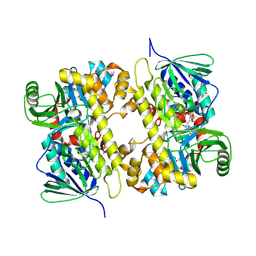 | | Crystal structure of MibH, a lathipeptide tryptophan 5-halogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Tryptophane-5-halogenase | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Two Flavoenzymes Catalyze the Post-Translational Generation of 5-Chlorotryptophan and 2-Aminovinyl-Cysteine during NAI-107 Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5X21
 
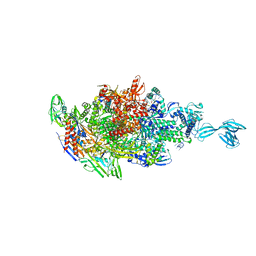 | | Crystal structure of Thermus thermophilus transcription initiation complex with GpA and pseudouridimycin (PUM) | | Descriptor: | (1S)-1,4-anhydro-5-[(N-carbamimidoylglycyl-N~2~-hydroxy-L-glutaminyl)amino]-5-deoxy-1-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-D-ribitol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zhang, Y, Ebright, R. | | Deposit date: | 2017-01-29 | | Release date: | 2017-07-05 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (3.323 Å) | | Cite: | Antibacterial Nucleoside-Analog Inhibitor of Bacterial RNA Polymerase.
Cell, 169, 2017
|
|
5X22
 
 | |
5EHK
 
 | |
