7L3P
 
 | |
7KP9
 
 | | asymmetric hTNF-alpha | | Descriptor: | 1-{[2-(difluoromethoxy)phenyl]methyl}-2-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-benzimidazole, Tumor necrosis factor | | Authors: | Arakaki, T.L, Abendroth, J, Fairman, J.W, Foley, A, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the disruption of TNF-TNFR1 signalling by small molecules stabilising a distorted TNF.
Nat Commun, 12, 2021
|
|
7L4G
 
 | |
7KQA
 
 | |
6CFP
 
 | |
7KN1
 
 | |
7L2A
 
 | |
7L3Q
 
 | |
7KQZ
 
 | |
6CXM
 
 | |
6CY1
 
 | |
7RY7
 
 | |
3P10
 
 | |
6CY5
 
 | |
7LXI
 
 | |
7S5E
 
 | |
7S87
 
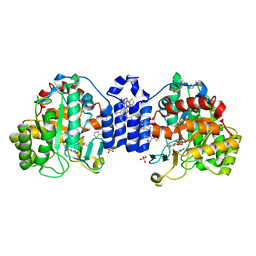 | | Crystal Structure of Dihydroorotate dehydrogenase from Plasmodium falciparum in complex with Orotate, FMN, and inhibitor NCGC00600348-01 | | Descriptor: | (4R)-3-methyl-5-[(4R)-4-methyl-3,4-dihydroisoquinolin-2(1H)-yl]thieno[2,3-e][1,2,4]triazolo[4,3-c]pyrimidine, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-09-17 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Dihydroorotate dehydrogenase from Plasmodium falciparum in complex with Orotate, FMN, and inhibitor NCGC00600348-01
To Be Published
|
|
7SBC
 
 | |
7RZO
 
 | |
7SIQ
 
 | |
6D8W
 
 | |
3P0Z
 
 | |
6D46
 
 | |
6D47
 
 | |
6DEG
 
 | |
