4V0S
 
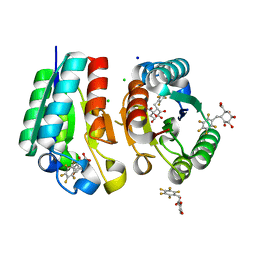 | | Crystal structure of Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3,4-DIHYDROXY-2-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]BENZOIC ACID, 3-DEHYDROQUINATE DEHYDRATASE, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Santiago, C, Lamb, H, Hawkins, A.R, Maneiro, M, Peon, A, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-09-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
4URH
 
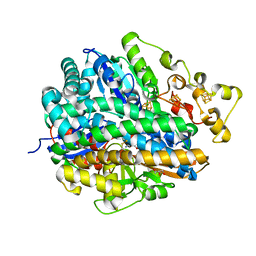 | | High-resolution structure of partially oxidized D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystallographic studies of [NiFe]-hydrogenase mutants: towards consensus structures for the elusive unready oxidized states.
J. Biol. Inorg. Chem., 20, 2015
|
|
4WWS
 
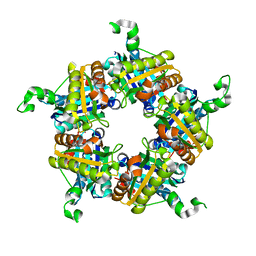 | |
8Q5P
 
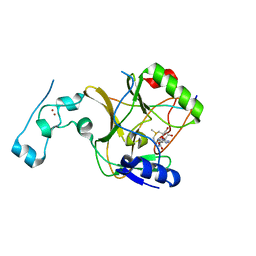 | | Structure of the lysine methyltransferase SETD2 in complex with a peptide derived from human tyrosine kinase ACK1 | | Descriptor: | Activated CDC42 kinase 1, Histone-lysine N-methyltransferase SETD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Mechaly, A, Le Coadou, L, Dupret, J.M, Haouz, A, Rodrigues Lima, F. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Structural and enzymatic evidence for the methylation of the ACK1 tyrosine kinase by the histone lysine methyltransferase SETD2.
Biochem.Biophys.Res.Commun., 695, 2024
|
|
109D
 
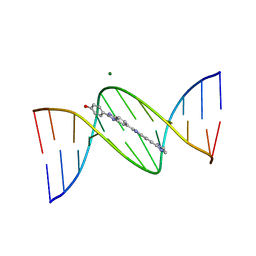 | | VARIABILITY IN DNA MINOR GROOVE WIDTH RECOGNISED BY LIGAND BINDING: THE CRYSTAL STRUCTURE OF A BIS-BENZIMIDAZOLE COMPOUND BOUND TO THE DNA DUPLEX D(CGCGAATTCGCG)2 | | Descriptor: | 5-(2-IMIDAZOLINYL)-2-[2-(4-HYDROXYPHENYL)-5-BENZIMIDAZOLYL]BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Czarny, A, Boykin, D.W, Wood, A.A, Nunn, C.M, Neidle, S, Zhao, M, Wilson, W.D. | | Deposit date: | 1995-02-15 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variability in DNA minor groove width recognised by ligand binding: the crystal structure of a bis-benzimidazole compound bound to the DNA duplex d(CGCGAATTCGCG)2.
Nucleic Acids Res., 23, 1995
|
|
1ACB
 
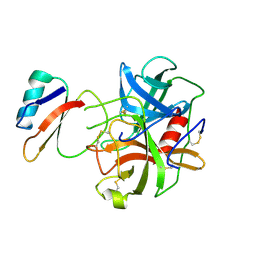 | | CRYSTAL AND MOLECULAR STRUCTURE OF THE BOVINE ALPHA-CHYMOTRYPSIN-EGLIN C COMPLEX AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-CHYMOTRYPSIN, Eglin C | | Authors: | Bolognesi, M, Frigerio, F, Coda, A, Pugliese, L, Lionetti, C, Menegatti, E, Amiconi, G, Schnebli, H.P, Ascenzi, P. | | Deposit date: | 1991-11-08 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal and molecular structure of the bovine alpha-chymotrypsin-eglin c complex at 2.0 A resolution.
J.Mol.Biol., 225, 1992
|
|
1A2C
 
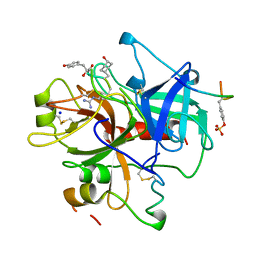 | | Structure of thrombin inhibited by AERUGINOSIN298-A from a BLUE-GREEN ALGA | | Descriptor: | Aeruginosin 298-A, Hirudin variant-2, SODIUM ION, ... | | Authors: | Rios-Steiner, J.L, Murakami, M, Tulinsky, A. | | Deposit date: | 1997-12-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Thrombin Inhibited by Aeruginosin 298-A from a Blue-Green Alga
J.Am.Chem.Soc., 120, 1998
|
|
1A0R
 
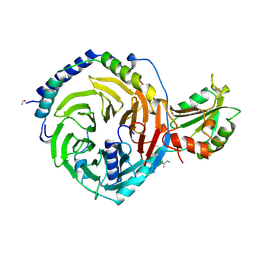 | | HETEROTRIMERIC COMPLEX OF PHOSDUCIN/TRANSDUCIN BETA-GAMMA | | Descriptor: | FARNESYL, PHOSDUCIN, TRANSDUCIN (BETA SUBUNIT), ... | | Authors: | Loew, A, Ho, Y.-K, Blundell, T.L, Bax, B. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosducin induces a structural change in transducin beta gamma.
Structure, 6, 1998
|
|
1A8G
 
 | | HIV-1 PROTEASE IN COMPLEX WITH SDZ283-910 | | Descriptor: | HIV-1 PROTEASE, benzyl [(1R)-1-({(1S,2S,3S)-1-benzyl-2-hydroxy-4-({(1S)-1-[(2-hydroxy-4-methoxybenzyl)carbamoyl]-2-methylpropyl}amino)-3-[(4-methoxybenzyl)amino]-4-oxobutyl}carbamoyl)-2,2-dimethylpropyl]carbamate | | Authors: | Kallen, J, Billich, A, Scholz, D, Auer, M, Kungl, A. | | Deposit date: | 1998-03-24 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure and conformational dynamics of the HIV-1 protease in complex with the inhibitor SDZ283-910: agreement of time-resolved spectroscopy and molecular dynamics simulations.
J.Mol.Biol., 286, 1999
|
|
1ABO
 
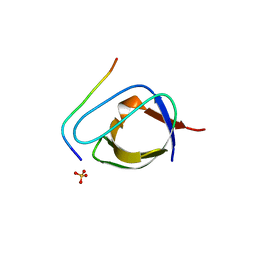 | | CRYSTAL STRUCTURE OF THE COMPLEX OF THE ABL TYROSINE KINASE SH3 DOMAIN WITH 3BP-1 SYNTHETIC PEPTIDE | | Descriptor: | 3BP-1 SYNTHETIC PEPTIDE, 10 RESIDUES, ABL TYROSINE KINASE, ... | | Authors: | Musacchio, A, Wilmanns, M, Saraste, M. | | Deposit date: | 1995-05-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
1AK8
 
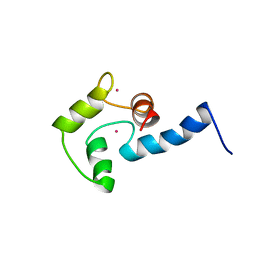 | | NMR SOLUTION STRUCTURE OF CERIUM-LOADED CALMODULIN AMINO-TERMINAL DOMAIN (CE2-TR1C), 23 STRUCTURES | | Descriptor: | CALMODULIN, CERIUM (III) ION | | Authors: | Bentrop, D, Bertini, I, Cremonini, M.A, Forsen, S, Luchinat, C, Malmendal, A. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the paramagnetic complex of the N-terminal domain of calmodulin with two Ce3+ ions by 1H NMR.
Biochemistry, 36, 1997
|
|
1AKT
 
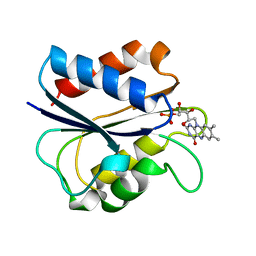 | | G61N OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1A5G
 
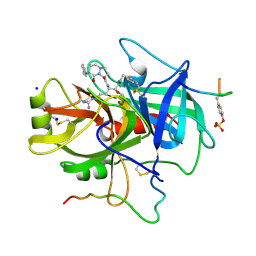 | | HUMAN THROMBIN COMPLEXED WITH NOVEL SYNTHETIC PEPTIDE MIMETIC INHIBITOR AND HIRUGEN | | Descriptor: | (1S,7S)-7-amino-7-benzyl-N-[(1S)-4-carbamimidamido-1-{(1S)-1-hydroxy-2-oxo-2-[(2-phenylethyl)amino]ethyl}butyl]-8-oxohexahydro-1H-pyrazolo[1,2-a]pyridazine-1-carboxamide, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | St Charles, R, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-02-16 | | Release date: | 1998-05-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
1AB1
 
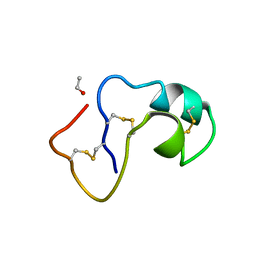 | | SI FORM CRAMBIN | | Descriptor: | CRAMBIN (SER22/ILE25), ETHANOL | | Authors: | Teeter, M.M, Yamano, A. | | Deposit date: | 1997-01-31 | | Release date: | 1997-08-12 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Crystal structure of Ser-22/Ile-25 form crambin confirms solvent, side chain substate correlations.
J.Biol.Chem., 272, 1997
|
|
1ABJ
 
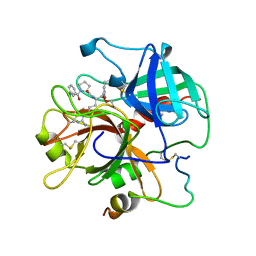 | |
1A0Z
 
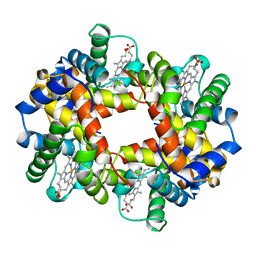 | | HEMOGLOBIN (VAL BETA1 MET) MUTANT | | Descriptor: | HEMOGLOBIN (ALPHA CHAIN), HEMOGLOBIN (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of human hemoglobin with mutations at tryptophan 37beta: structural basis for a high-affinity T-state,.
Biochemistry, 37, 1998
|
|
1AKR
 
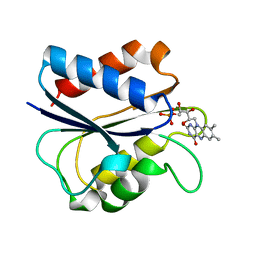 | | G61A OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1AKW
 
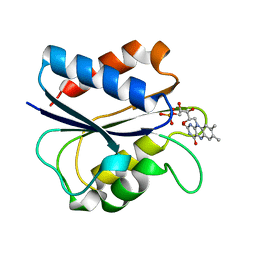 | | G61L OXIDIZED FLAVODOXIN MUTANT | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Mccarthy, A, Walsh, M, Higgins, T. | | Deposit date: | 1997-05-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: thermodynamic properties and crystal structures of glycine-61 mutants.
Biochemistry, 37, 1998
|
|
1A6S
 
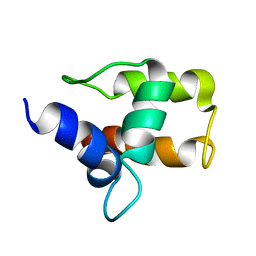 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
1ABI
 
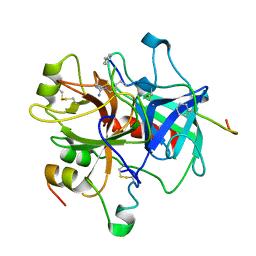 | |
1ABQ
 
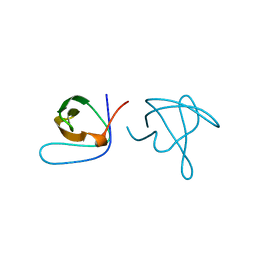 | |
1AMY
 
 | |
1B5G
 
 | | HUMAN THROMBIN COMPLEXED WITH NOVEL SYNTHETIC PEPTIDE MIMETIC INHIBITOR AND HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, SODIUM ION, ... | | Authors: | St Charles, R, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
1AZF
 
 | | CHICKEN EGG WHITE LYSOZYME CRYSTAL GROWN IN BROMIDE SOLUTION | | Descriptor: | BROMIDE ION, LYSOZYME | | Authors: | Lim, K, Nadarajah, A, Forsythe, E.L, Pusey, M.L. | | Deposit date: | 1997-11-17 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Locations of bromide ions in tetragonal lysozyme crystals.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AVD
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE TETRAGONAL CRYSTAL FORM OF EGG-WHITE AVIDIN IN ITS FUNCTIONAL COMPLEX WITH BIOTIN AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AVIDIN, BIOTIN | | Authors: | Pugliese, L, Coda, A, Malcovati, M, Bolognesi, M. | | Deposit date: | 1993-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of the tetragonal crystal form of egg-white avidin in its functional complex with biotin at 2.7 A resolution.
J.Mol.Biol., 231, 1993
|
|
