2QXM
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed to Burned Meat Compound PhIP | | Descriptor: | 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Gil, G, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-08-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2QGT
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed to an Ether Estradiol Compound | | Descriptor: | (9BETA,11ALPHA,13ALPHA,14BETA,17ALPHA)-11-(METHOXYMETHYL)ESTRA-1(10),2,4-TRIENE-3,17-DIOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2QGW
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed with a Chloro-Indazole Compound | | Descriptor: | 3-CHLORO-2-(4-HYDROXYPHENYL)-2H-INDAZOL-5-OL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2QA8
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Mutant 537S Complexed with Genistein | | Descriptor: | Estrogen receptor, GENISTEIN, nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-14 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
7C4U
 
 | | MicroED structure of orthorhombic Vancomycin at 1.2 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7C4V
 
 | | MicroED structure of anorthic Vancomycin at 1.05 A resolution | | Descriptor: | CHLORIDE ION, Vancomycin, vancosamine-(1-2)-beta-D-glucopyranose | | Authors: | Fan, Q, Zhou, H, Li, X, Wang, J. | | Deposit date: | 2020-05-18 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Precise Control Over Kinetics of Molecular Assembly: Production of Particles with Tunable Sizes and Crystalline Forms.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8I1Y
 
 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I2J
 
 | |
8I1Z
 
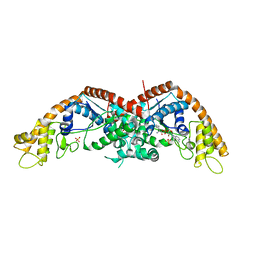 | | E. coli tryptophanyl-tRNA synthetase bound with a chemical fragment | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)ethanone, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
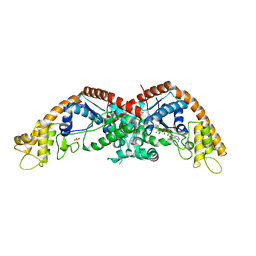
8I2C
 
 | |
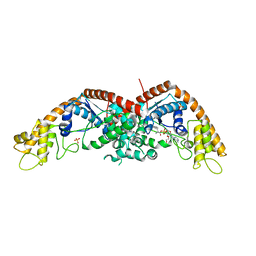
8I2A
 
 | |
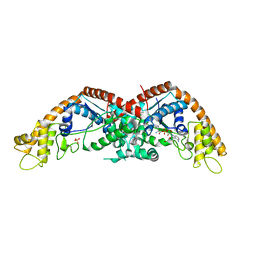
8I27
 
 | |
8I2L
 
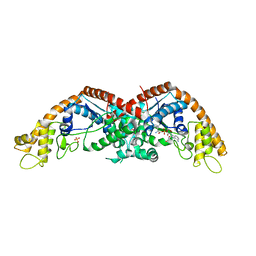 | |
8I1W
 
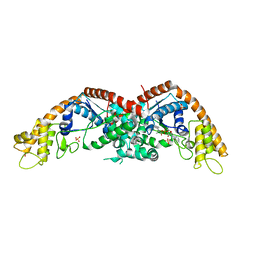 | |
8I2M
 
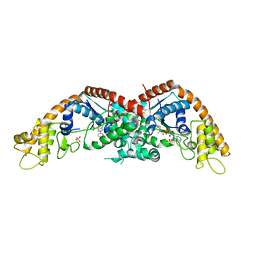 | |
8I4I
 
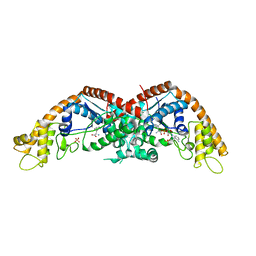 | |
4L36
 
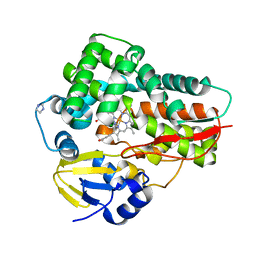 | | Crystal structure of the cytochrome P450 enzyme TxtE | | Descriptor: | HEME B/C, IMIDAZOLE, Putative P450-like protein | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Wang, Z.J, Zhou, H, Yang, M, Chen, Y.X, Tang, L, He, J.H. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Insights into the Mechanism for Recognizing Substrate of the Cytochrome P450 Enzyme TxtE.
Plos One, 8, 2013
|
|
5TGZ
 
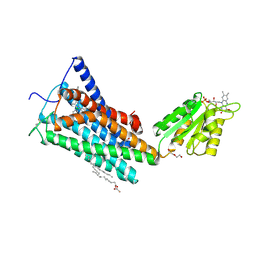 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
7TIL
 
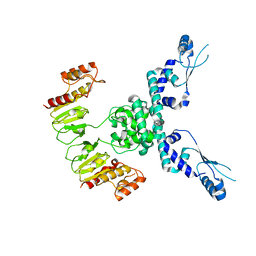 | | CryoEM structure of JetD from Pseudomonas aeruginosa | | Descriptor: | JetD | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-01-13 | | Release date: | 2022-10-05 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
7TQD
 
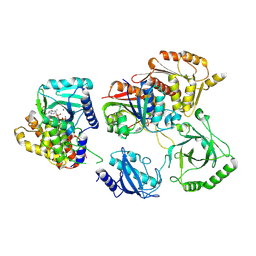 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TO3
 
 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TSX
 
 | | Structure of Enterobacter cloacae Cap2 bound to CdnD02 C-terminus, Apo state | | Descriptor: | Cap2, Cyclic AMP-AMP-GMP synthase | | Authors: | Ye, Q, Gu, Y, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7TSQ
 
 | | Structure of Enterobacter cloacae Cap2 bound to CdnD02 C-terminus, AMP state | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cap2, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Ye, Q, Gu, Y, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
7VRC
 
 | |
5X4J
 
 | | The crystal structure of Pyrococcus furiosus RecJ (Zn-soaking) | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Li, M.J, Yi, G.S, Yu, F, Zhou, H, Chen, J.N, Xu, C.Y, Wang, F.P, Xiao, X, He, J.H, Liu, X.P. | | Deposit date: | 2017-02-13 | | Release date: | 2018-02-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45.
Nucleic Acids Res., 45, 2017
|
|
