6LJM
 
 | |
6LJN
 
 | |
6LJK
 
 | |
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | 分子名称: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | 著者 | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | 登録日 | 2022-03-31 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
8GUO
 
 | |
8GUP
 
 | |
8GUN
 
 | |
7FAQ
 
 | | Crystal structure of PDE5A in complex with inhibitor L1 | | 分子名称: | 2-[(4-chlorophenyl)methyl]-5-methoxy-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Wu, D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.200136 Å) | | 主引用文献 | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
7FAR
 
 | | Crystal structure of PDE5A in complex with inhibitor L12 | | 分子名称: | 5-[bis(fluoranyl)methoxy]-2-[(4-chlorophenyl)methyl]-10-(3-methoxypropyl)-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, ZINC ION, ... | | 著者 | Wu, D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.40006471 Å) | | 主引用文献 | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
7Y73
 
 | |
7Y8I
 
 | | Crystal structure of sDscam FNIII3 domain, isoform alpha7 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dscam, ... | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y9A
 
 | | Crystal structure of sDscam Ig1-2 domains, isoform beta2v6 | | 分子名称: | Down Syndrome Cell Adhesion Molecules, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)][beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y5J
 
 | |
7Y8S
 
 | |
7Y6O
 
 | |
7Y54
 
 | |
7Y5R
 
 | |
7Y4X
 
 | |
7Y6E
 
 | |
7Y95
 
 | | Crystal structure of sDscam Ig1 domain, isoform beta6v2 | | 分子名称: | Dscam, GLYCEROL, SODIUM ION | | 著者 | Chen, Q, Yu, Y, Cheng, J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y8H
 
 | |
8HN9
 
 | | Human SIRT3 Recognizing CCNE2K348la peptide | | 分子名称: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | 著者 | Wang, Y, Ding, W. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
7VS2
 
 | |
7X8C
 
 | | Crystal structure of a KTSC family protein from Euryarchaeon Methanolobus vulcani | | 分子名称: | KTSC domain-containing protein, SODIUM ION | | 著者 | Zhang, Z.F, Zhu, K.L, Chen, Y.Y, Cao, P, Gong, Y. | | 登録日 | 2022-03-12 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Biochemical and structural characterization of a KTSC family single-stranded DNA-binding protein from Euryarchaea.
Int.J.Biol.Macromol., 216, 2022
|
|
7YI4
 
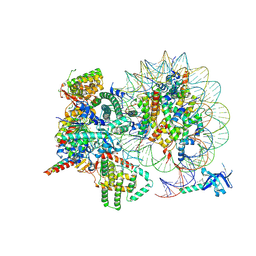 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | 分子名称: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
