7YMH
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to noradrenaline | | Descriptor: | Nb29, Noradrenaline, alpha1A-adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
2MDJ
 
 | |
7YM8
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to oxymetazoline | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Oxymetazoline, alpha1A adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
2MDC
 
 | | Solution structure of the WW domain of HYPB | | Descriptor: | Histone-lysine N-methyltransferase SETD2 | | Authors: | Gao, Y.G. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin
Structure, 22, 2014
|
|
7YUB
 
 | | S1P-bound human SPNS2 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, NbFab-H-chain, NbFab-L-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUF
 
 | | apo human SPNS2 | | Descriptor: | NbFab H-chain, NbFab L-chain, Sphingosine-1-phosphate transporter SPNS2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUD
 
 | | FTY720p-bound human SPNS2 | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, NbFab L-chain, NbFab-H-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7Y53
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
2MDI
 
 | | Solution structure of the PP2WW mutant (KPP2WW) of HYPB | | Descriptor: | Histone-lysine N-methyltransferase SETD2 | | Authors: | Gao, Y.G, Hu, H.Y. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin.
Structure, 22, 2014
|
|
7WK0
 
 | | Local refine of Omicron spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJZ
 
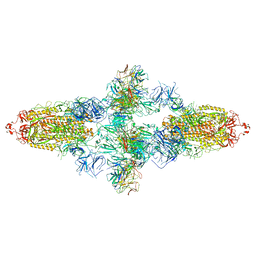 | | Omicron Spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7EMN
 
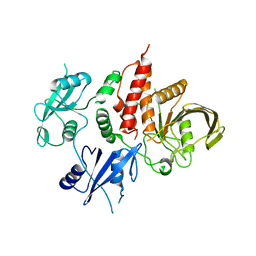 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
8IVQ
 
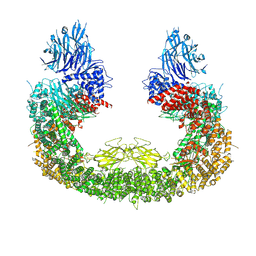 | | Cryo-EM structure of mouse BIRC6, Global map | | Descriptor: | Isoform 2 of Baculoviral IAP repeat-containing protein 6 | | Authors: | Liu, S, Jiang, T, Bu, F, Zhao, J, Wang, G, Li, N, Gao, N, Qiu, X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy.
Nat Commun, 15, 2024
|
|
7FCP
 
 | |
7X2H
 
 | |
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 (3.2A) | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
4KXJ
 
 | | Crystal structure of HCoV-OC43 N-NTD complexed with PJ34 | | Descriptor: | Nucleoprotein, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | Authors: | Lin, S.Y, Hou, M.H. | | Deposit date: | 2013-05-27 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target
J.Med.Chem., 57, 2014
|
|
4LM7
 
 | |
4LI4
 
 | |
4LM9
 
 | |
4LMC
 
 | |
