1P9E
 
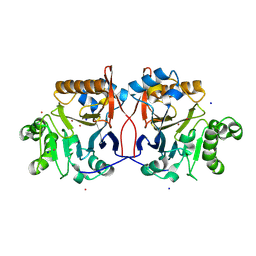 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | 分子名称: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | 著者 | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | 登録日 | 2003-05-11 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
4GYV
 
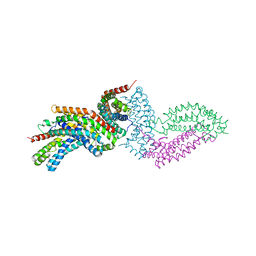 | | Crystal structure of the DH domain of FARP2 | | 分子名称: | FERM, RhoGEF and pleckstrin domain-containing protein 2 | | 著者 | He, X, Zhang, X. | | 登録日 | 2012-09-05 | | 公開日 | 2013-03-13 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural Basis for Autoinhibition of the Guanine Nucleotide Exchange Factor FARP2.
Structure, 21, 2013
|
|
2H3B
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor 1 | | 分子名称: | Nicotinamide phosphoribosyltransferase, SULFATE ION | | 著者 | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | 登録日 | 2006-05-22 | | 公開日 | 2006-06-20 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2GZY
 
 | |
4GZN
 
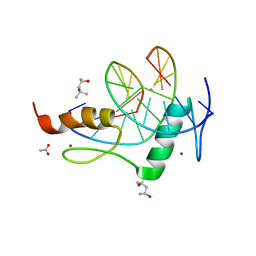 | | Mouse ZFP57 zinc fingers in complex with methylated DNA | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Liu, Y, Zhang, X, Cheng, X. | | 登録日 | 2012-09-06 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence.
Genes Dev., 26, 2012
|
|
4GZU
 
 | |
2GZZ
 
 | |
2H3D
 
 | | Crystal Structure of Mouse Nicotinamide Phosphoribosyltransferase/Visfatin/Pre-B Cell Colony Enhancing Factor in Complex with Nicotinamide Mononuleotide | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase | | 著者 | Wang, T, Zhang, X, Bheda, P, Revollo, J.R, Imai, S.I, Wolberger, C. | | 登録日 | 2006-05-22 | | 公開日 | 2006-06-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of Nampt/PBEF/visfatin, a mammalian NAD(+) biosynthetic enzyme.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2NYD
 
 | |
4ZDS
 
 | | Crystal Structure of core DNA binding domain of Arabidopsis Thaliana Transcription Factor Ethylene-Insensitive 3 | | 分子名称: | Protein ETHYLENE INSENSITIVE 3 | | 著者 | Song, J, Zhu, C, Zhang, X, Wen, X, Liu, L, Peng, J, Guo, H, Yi, C. | | 登録日 | 2015-04-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Biochemical and Structural Insights into the Mechanism of DNA Recognition by Arabidopsis ETHYLENE INSENSITIVE3.
Plos One, 10, 2015
|
|
5K9N
 
 | | Structural and Mechanistic Analysis of Drosophila melanogaster Polyamine N acetyltransferase, an enzyme that Catalyzes the Formation of N acetylagmatine | | 分子名称: | Polyamine N acetyltransferase | | 著者 | Dempsey, D.R, Nichols, D.A, Battistini, M.R, Pemberton, O, Ospina, S.R, Zhang, X, Carpenter, A.-M, Chen, Y, Merkler, D.J. | | 登録日 | 2016-06-01 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Mechanistic Analysis of Drosophila melanogaster Agmatine N-Acetyltransferase, an Enzyme that Catalyzes the Formation of N-Acetylagmatine.
Sci Rep, 7, 2017
|
|
7SII
 
 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | 分子名称: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | 著者 | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | 登録日 | 2021-10-14 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
5CG9
 
 | | NgTET1 in complex with 5mC DNA in space group P3221 | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DNA (5'-D(*TP*GP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | 登録日 | 2015-07-09 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.693 Å) | | 主引用文献 | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
5CG8
 
 | | NgTET1 in complex with 5hmC DNA | | 分子名称: | 2-OXOGLUTARIC ACID, DNA (5'-D(*AP*GP*AP*AP*TP*TP*CP*CP*GP*TP*TP*CP*CP*A)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*(5HC)P*GP*GP*AP*AP*TP*TP*CP*T)-3'), ... | | 著者 | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | 登録日 | 2015-07-09 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-13 | | 公開日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-14 | | 公開日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | 分子名称: | Insulin fusion, Insulin receptor | | 著者 | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | 登録日 | 2020-04-14 | | 公開日 | 2021-04-14 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7CCR
 
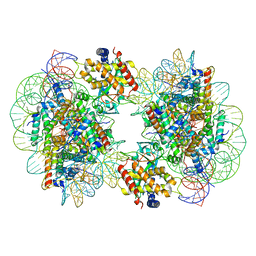 | | Structure of the 2:2 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7CCQ
 
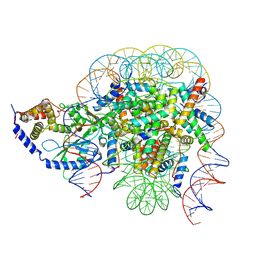 | | Structure of the 1:1 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | 著者 | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | 登録日 | 2020-06-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7UDC
 
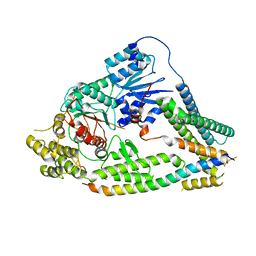 | | cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex class1 | | 分子名称: | Synaptobrevin-2, Syntaxin-1A, Syntaxin-binding protein 1 | | 著者 | Rizo, J, Bai, X, Stepien, K.P, Xu, J, Zhang, X. | | 登録日 | 2022-03-18 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex.
Sci Adv, 8, 2022
|
|
7UDB
 
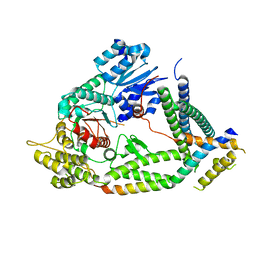 | | Cryo-EM structure of a synaptobrevin-Munc18-1-syntaxin-1 complex class 2 | | 分子名称: | Synaptobrevin-2, Syntaxin-1A, Syntaxin-binding protein 1 | | 著者 | Rizo, J, Bai, X, Stepien, K.P, Xu, J, Zhang, X. | | 登録日 | 2022-03-18 | | 公開日 | 2022-04-06 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | SNARE assembly enlightened by cryo-EM structures of a synaptobrevin-Munc18-1-syntaxin-1 complex.
Sci Adv, 8, 2022
|
|
