4Z2G
 
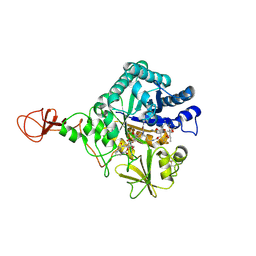 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 26 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {4-[N'-(methylcarbamoyl)carbamimidamido]butyl}carbamate, Chitinase B | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2L
 
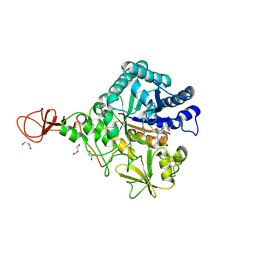 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 33 | | Descriptor: | (3R,4S,5S,6R,7R,9R,10S,11S,12R,13S,14R)-14-ethyl-7,10,12,13-tetrahydroxy-3,5,7,9,11,13-hexamethyl-2-oxo-6-(prop-2-yn-1-yloxy)oxacyclotetradecan-4-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2J
 
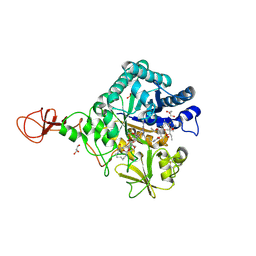 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 31 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {5-[N'-(methylcarbamoyl)carbamimidamido]pentyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2K
 
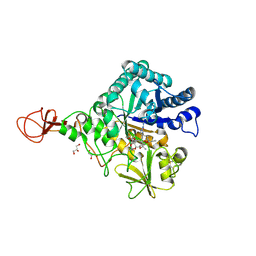 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 32 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {7-[N'-(methylcarbamoyl)carbamimidamido]heptyl}carbamate, Chitinase B, GLYCEROL | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2H
 
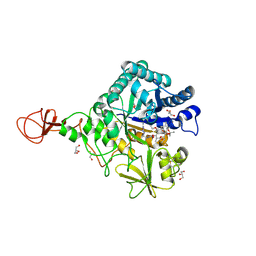 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 29 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {2-[N'-(methylcarbamoyl)carbamimidamido]ethyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2I
 
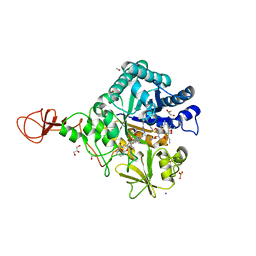 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 30 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, Chitinase B, GLYCEROL, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
2KUQ
 
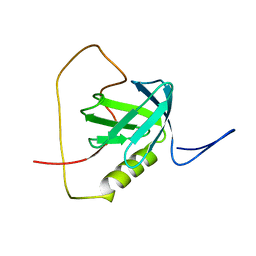 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | Fibroblast growth factor receptor substrate 3,LINKER,ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2KUP
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of HALK | | Descriptor: | 19-residue peptide from ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
5X9O
 
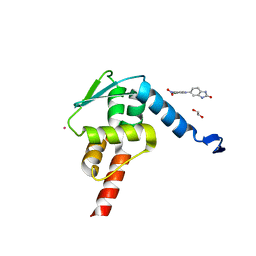 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
1DHY
 
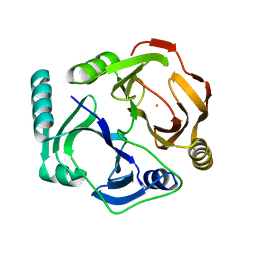 | | KKS102 BPHC ENZYME | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, FE (III) ION | | Authors: | Senda, T, Sugiyama, K, Narita, H, Mitsui, Y. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. strain KKS102.
J.Mol.Biol., 255, 1996
|
|
5URM
 
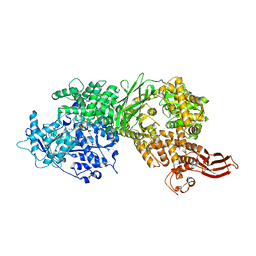 | | Crystal structure of human BRR2 in complex with T-1206548 | | Descriptor: | 3-(5-{[(2R)-5-amino-2-cyclohexyl-7-oxo-2,3-dihydro-7H-[1,3,4]thiadiazolo[3,2-a]pyrimidin-6-yl]methyl}furan-2-yl)benzoic acid, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-11 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URJ
 
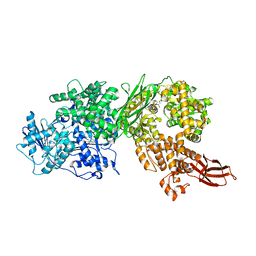 | | Crystal structure of human BRR2 in complex with T-3905516 | | Descriptor: | 6-benzyl-3-[(2R)-2-(3-fluoropyridin-2-yl)-6-methyl-3,4-dihydro-2H-1-benzopyran-7-yl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(3H,8H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Klein, M.G, Tjhen, R, Qin, L. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5URK
 
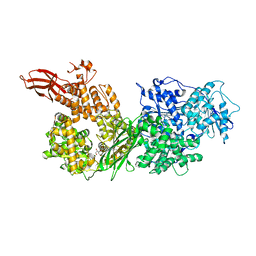 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
5B6F
 
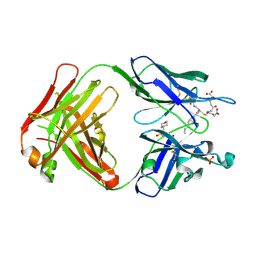 | | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4 | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-(2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Sugahara, M, Ago, H, Saino, H, Miyano, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Fab fragment of an anti-Leukotriene C4 monoclonal antibody complexed with LTC4
To Be Published
|
|
2YS5
 
 | | Solution structure of the complex of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | ALK tyrosine kinase receptor, Fibroblast growth factor receptor substrate 3 | | Authors: | Li, H, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
2YT2
 
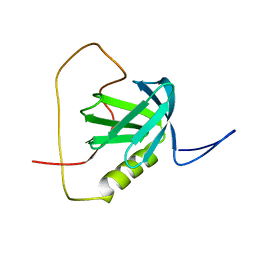 | | Solution structure of the chimera of the PTB domain of SNT-2 and 19-residue peptide (aa 1571-1589) of hALK | | Descriptor: | Fibroblast growth factor receptor substrate 3 and ALK tyrosine kinase receptor | | Authors: | Li, H, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2014-02-12 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of nucleophosmin-anaplastic lymphoma kinase oncoprotein by the phosphotyrosine binding domain of Suc1-associated neurotrophic factor-induced tyrosine-phosphorylated target-2
J.Struct.Funct.Genom., 11, 2010
|
|
7DC8
 
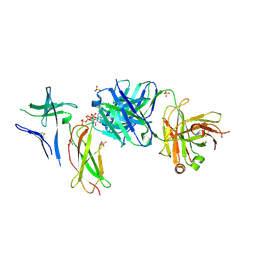 | | Crystal structure of Switch Ab Fab and hIL6R in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interleukin-6 receptor subunit alpha, SULFATE ION, ... | | Authors: | Kadono, S, Fukami, T.A, Kawauchi, H, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.757 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
7DC7
 
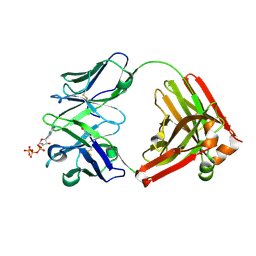 | | Crystal structure of D12 Fab-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D12 Fab heavy chain, D12 Fab light chain | | Authors: | Kawauchi, H, Fukami, T.A, Tatsumi, K, Torizawa, T, Mimoto, F. | | Deposit date: | 2020-10-23 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploitation of Elevated Extracellular ATP to Specifically Direct Antibody to Tumor Microenvironment.
Cell Rep, 33, 2020
|
|
2ZTW
 
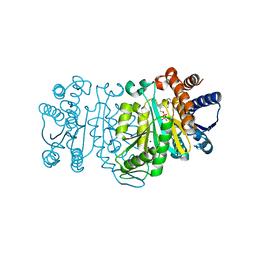 | | Structure of 3-isopropylmalate dehydrogenase in complex with the inhibitor and NAD+ | | Descriptor: | (2Z)-2-hydroxy-3-(methylsulfanyl)prop-2-enoic acid, 3-isopropylmalate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Nango, E, Kumasaka, T, Eguchi, T. | | Deposit date: | 2008-10-10 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of 3-isopropylmalate dehydrogenase in complex with NAD(+) and a designed inhibitor
Bioorg.Med.Chem., 17, 2009
|
|
1EIL
 
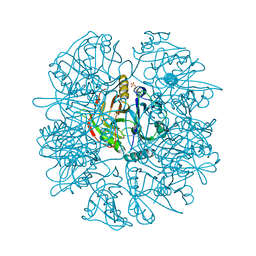 | | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, FE (III) ION, SULFATE ION | | Authors: | Senda, T. | | Deposit date: | 2000-02-26 | | Release date: | 2001-02-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of substrate free and complex forms of reactivated BphC, an extradiol type ring-cleavage dioxygenase.
J.Inorg.Biochem., 83, 2001
|
|
1EIR
 
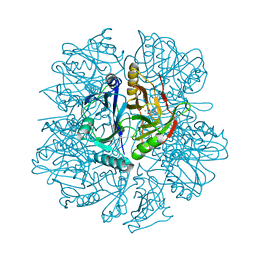 | | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE | | Descriptor: | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE, BIPHENYL-2,3-DIOL, FE (III) ION | | Authors: | Senda, T. | | Deposit date: | 2000-02-28 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of substrate free and complex forms of reactivated BphC, an extradiol type ring-cleavage dioxygenase.
J.Inorg.Biochem., 83, 2001
|
|
3ASJ
 
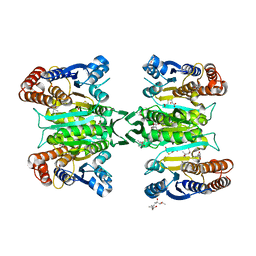 | | Crystal structure of homoisocitrate dehydrogenase in complex with a designed inhibitor | | Descriptor: | (2Z)-3-[(carboxymethyl)sulfanyl]-2-hydroxyprop-2-enoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[(carboxymethyl)sulfanyl]-2-oxopropanoic acid, ... | | Authors: | Nango, E, Kumasaka, T, Eguchi, T. | | Deposit date: | 2010-12-13 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermus thermophilus homoisocitrate dehydrogenase in complex with a designed inhibitor
J.Biochem., 150, 2011
|
|
2YZE
 
 | | Crystal structure of uricase from Arthrobacter globiformis | | Descriptor: | (dihydroxyboranyloxy-hydroxy-boranyl)oxylithium, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
2YZD
 
 | | Crystal structure of uricase from Arthrobacter globiformis in complex with 8-azaxanthin (inhibitor) | | Descriptor: | 8-AZAXANTHINE, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
2YZB
 
 | | Crystal structure of uricase from Arthrobacter globiformis in complex with uric acid (substrate) | | Descriptor: | URIC ACID, Uricase | | Authors: | Juan, E.C.M, Hossain, M.T, Hoque, M.M, Suzuki, K, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2007-05-05 | | Release date: | 2008-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping of the uric acid substrate in the crystal structure of urate oxidase from Arthrobacter globiformis
To be Published
|
|
