1QZ7
 
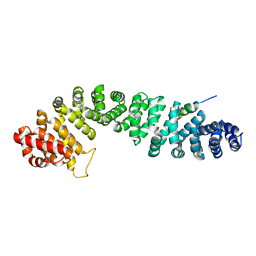 | | Beta-catenin binding domain of Axin in complex with beta-catenin | | Descriptor: | Axin, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Kimelman, D, Xu, W. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a beta-catenin/Axin complex suggests a mechanism for the {beta}-catenin destruction complex
GENES DEV., 17, 2003
|
|
1TH1
 
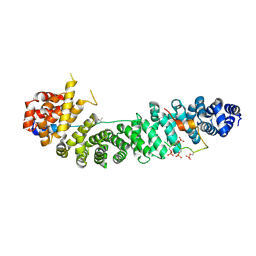 | | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Le Trong, I, Hinds, T.R, Stenkamp, R, Kimelman, D, Xu, W. | | Deposit date: | 2004-05-31 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a beta-Catenin/APC Complex Reveals a Critical Role for APC Phosphorylation in APC Function.
Mol.Cell, 15, 2004
|
|
3M89
 
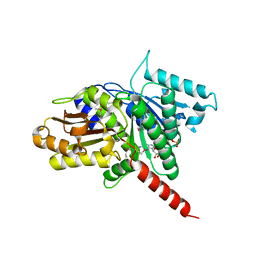 | | Structure of TubZ-GTP-g-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, FtsZ/tubulin-related protein | | Authors: | Ni, L, Xu, W, Schumacher, M.A. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3EZ2
 
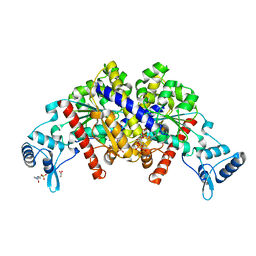 | | Partition protein-ADP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Schumacher, M.A, Dunham, T.D, Xu, W, Funnell, B. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
3MKW
 
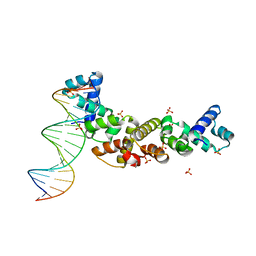 | | Structure of sopB(155-272)-18mer complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3MKY
 
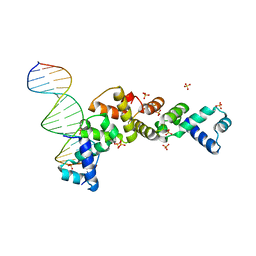 | | Structure of SopB(155-323)-18mer DNA complex, I23 form | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB, SULFATE ION | | Authors: | Schumacher, M.A, Piro, K, Xu, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
1YAX
 
 | |
3JS6
 
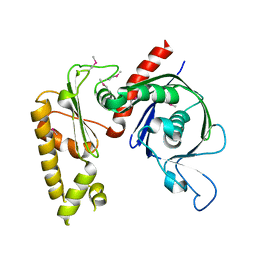 | | Crystal structure of apo psk41 parM protein | | Descriptor: | Uncharacterized ParM protein | | Authors: | Schumacher, M.A, Xu, W, Firth, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation.
J.Biol.Chem., 285, 2010
|
|
2IAE
 
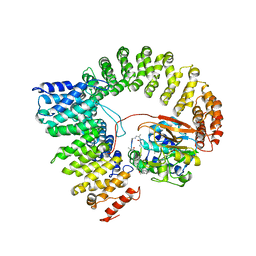 | | Crystal structure of a protein phosphatase 2A (PP2A) holoenzyme. | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Cho, U.S, Xu, W. | | Deposit date: | 2006-09-07 | | Release date: | 2006-12-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme.
Nature, 445, 2007
|
|
1XTN
 
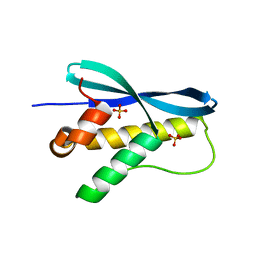 | | crystal structure of CISK-PX domain with sulfates | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase Sgk3 | | Authors: | Xing, Y, Liu, D, Zhang, R, Joachimiak, A, Songyang, Z, Xu, W. | | Deposit date: | 2004-10-22 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX)
J.Biol.Chem., 279, 2004
|
|
1XTE
 
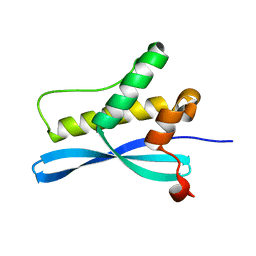 | | crystal structure of CISK-PX domain | | Descriptor: | Serine/threonine-protein kinase Sgk3 | | Authors: | Xing, Y, Liu, D, Zhang, R, Joachimiak, A, Songyang, Z, Xu, W. | | Deposit date: | 2004-10-21 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX)
J.Biol.Chem., 279, 2004
|
|
2KF7
 
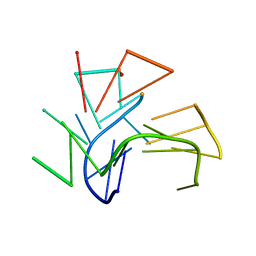 | | Structure of a two-G-tetrad basket-type intramolecular G-quadruplex formed by human telomeric repeats in K+ solution (with G7-to-BRG substitution) | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Lim, K.W, Amrane, S, Bouaziz, S, Xu, W, Mu, Y, Patel, D.J, Luu, K.N, Phan, A.T. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers
J.Am.Chem.Soc., 131, 2009
|
|
2KF8
 
 | | Structure of a two-G-tetrad basket-type intramolecular G-quadruplex formed by human telomeric repeats in K+ solution | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Lim, K.W, Amrane, S, Bouaziz, S, Xu, W, Mu, Y, Patel, D.J, Luu, K.N, Phan, A.T. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers
J.Am.Chem.Soc., 131, 2009
|
|
7XMN
 
 | | Structure of SARS-CoV-2 ORF8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein, ... | | Authors: | Chen, X, Xu, W. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylated, Lipid-Binding, CDR-Like Domains of SARS-CoV-2 ORF8 Indicate Unique Sites of Immune Regulation.
Microbiol Spectr, 11, 2023
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M60
 
 | | Crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Olmos Jr, J.L, Martinez III, E, Wang, F, Helmich, K.E, Singh, S, Xu, W, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-04 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4M7P
 
 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4N9Y
 
 | |
4NA0
 
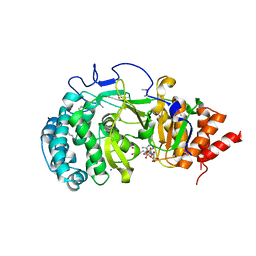 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4NA4
 
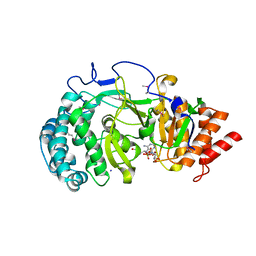 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2014-09-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4NA5
 
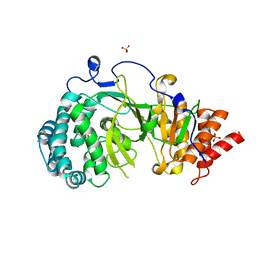 | |
4NA6
 
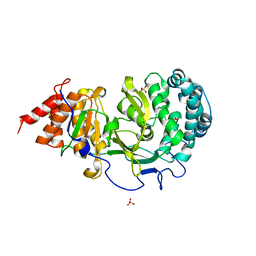 | |
4N9Z
 
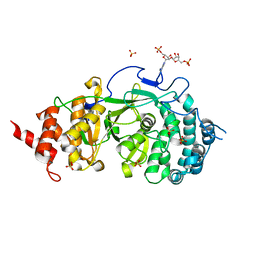 | |
4PIW
 
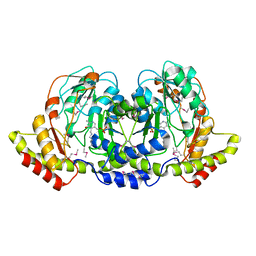 | | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12 | | Descriptor: | TDP-4-keto-6-deoxy-D-glucose transaminase family protein | | Authors: | Wang, F, Xu, W, Helmich, K.E, Singh, S, Yennamalli, R.M, Miller, M.D, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of sugar aminotransferase WecE from Escherichia coli K-12
To Be Published
|
|
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
