4ZRD
 
 | | Crystal structure of SMG1 F278N mutant | | Descriptor: | GLYCEROL, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
4ZRE
 
 | | Crystal structure of SMG1 F278D mutant | | Descriptor: | CHLORIDE ION, LIP1, secretory lipase (Family 3), ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-05-12 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of product-bound SMG1 lipase: active site gating implications.
Febs J., 282, 2015
|
|
7PAD
 
 | | The crystal structure of DW-0254 in complex with PDE6D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
7PAC
 
 | | The crystal structure of PDE6D in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
7PAE
 
 | | The crystal structure of Deltarasin in complex with PDE6D | | Descriptor: | ACETIC ACID, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, deltarasin | | Authors: | Beaumont, E, Williams, D. | | Deposit date: | 2021-07-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Validation of a small molecule inhibitor of PDE6D-RAS interaction with favorable anti-leukemic effects.
Blood Cancer J, 12, 2022
|
|
7QV2
 
 | | Bacillus subtilis collided disome (Collided 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
7QV3
 
 | |
7QV1
 
 | | Bacillus subtilis collided disome (Leading 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
8DH2
 
 | |
8DH1
 
 | | T7 RNA polymerase elongation complex with unnatural base dDs-PaTP pair | | Descriptor: | 1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-ribofuranosyl}-1H-pyrrole-2-carbaldehyde, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2022-06-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase.
Nat Commun, 14, 2023
|
|
8DH4
 
 | | T7 RNA polymerase elongation complex with unnatural base dPa-DsTP pair | | Descriptor: | (7P)-3-{5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-ribofuranosyl}-7-(thiophen-2-yl)-3H-imidazo[4,5-b]pyridine, MAGNESIUM ION, Non-template strand DNA, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2022-06-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase.
Nat Commun, 14, 2023
|
|
8DH3
 
 | | T7 RNA polymerase elongation complex with unnatural base dPa | | Descriptor: | Non-template strand DNA, RNA, T7 RNA polymerase, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2022-06-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase.
Nat Commun, 14, 2023
|
|
8DH5
 
 | |
8DH0
 
 | | T7 RNA polymerase elongation complex with unnatural base dDs | | Descriptor: | GLYCEROL, Non-template strand DNA, RNA, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2022-06-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription recognition of a hydrophobic unnatural base pair by T7 RNA polymerase.
Nat Commun, 14, 2023
|
|
2X6L
 
 | |
2X69
 
 | |
2X6G
 
 | |
2ZMV
 
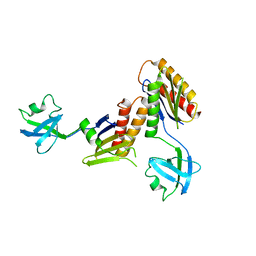 | | Crystal structure of Synbindin | | Descriptor: | Trafficking protein particle complex subunit 4 | | Authors: | Fan, F. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human synbindin reveals two conformations of longin domain
Biochem.Biophys.Res.Commun., 378, 2009
|
|
7WQ4
 
 | | Galanin-bound galanin receptor 2 in complex with Gq | | Descriptor: | CHOLESTEROL, Engineered Guanine nucleotide-binding protein G(q) subunit alpha, Galanin, ... | | Authors: | Duan, J, Shen, D.D, Xu, H.E, Zhang, Y, Jiang, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis for allosteric agonism and G protein subtype selectivity of galanin receptors
Nat Commun, 13, 2022
|
|
7WQ3
 
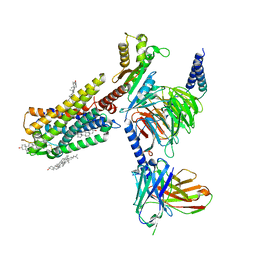 | | Galanin-bound galanin receptor 1 in complex with Gi | | Descriptor: | CHOLESTEROL, Galanin, Galanin receptor type 1, ... | | Authors: | Duan, J, Shen, D.D, Xu, H.E, Zhang, Y, Jiang, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-20 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for allosteric agonism and G protein subtype selectivity of galanin receptors
Nat Commun, 13, 2022
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
7YK7
 
 | | Cryo-EM structure of the DC591053-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Xu, H.E, Yang, D.H, Liu, H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YK6
 
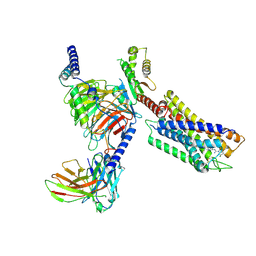 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YJ4
 
 | | Cryo-EM structure of the INSL5-bound human relaxin family peptidereceptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-19 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7DUQ
 
 | | Cryo-EM structure of the compound 2 and GLP-1-bound human GLP-1 receptor-Gs complex | | Descriptor: | CHOLESTEROL, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Cong, Z, Chen, L, Ma, H, Zhou, Q, Zou, X, Ye, C, Dai, A, Liu, Q, Huang, W, Sun, X, Wang, X, Xu, P, Zhao, L, Xia, T, Zhong, W, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
