4YPH
 
 | | Crystal Structure of MutY bound to its anti-substrate with the disulfide cross-linker reduced | | Descriptor: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(TP*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | Authors: | Wang, L, Lee, S, Verdine, G.L. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
1Y2M
 
 | | Crystal structure of phenylalanine ammonia-lyase from yeast Rhododporidium toruloides | | Descriptor: | Phenylalanine ammonia-lyase | | Authors: | Wang, L, Gamez, A, Sarkissian, C.N, Straub, M, Patch, M.G, Han, G.W, Scriver, C.R, Stevens, R.C. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based chemical modification strategy for enzyme replacement treatment of phenylketonuria.
Mol.Genet.Metab., 86, 2005
|
|
8UUK
 
 | | Pendrin in apo | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
4GA7
 
 | | Crystal structure of human serpinB1 mutant | | Descriptor: | Leukocyte elastase inhibitor | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4GAW
 
 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
6WLZ
 
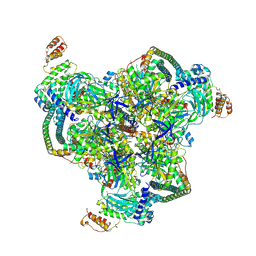 | | The V1 region of human V-ATPase in state 1 (focused refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SidK, V-type proton ATPase catalytic subunit A, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WM4
 
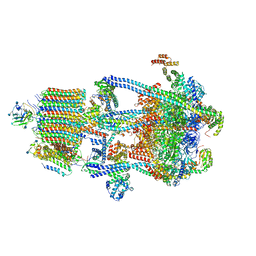 | | Human V-ATPase in state 3 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WLW
 
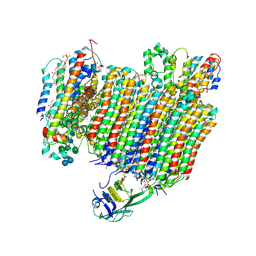 | | The Vo region of human V-ATPase in state 1 (focused refinement) | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, L, Wu, H, Fu, T.-M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WM2
 
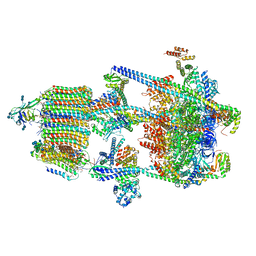 | | Human V-ATPase in state 1 with SidK and ADP | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WM3
 
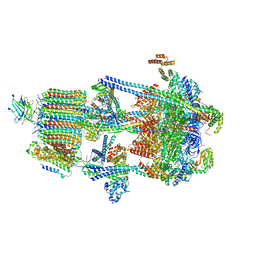 | | Human V-ATPase in state 2 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
4NSC
 
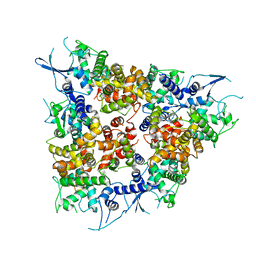 | | Crystal Structure of CBARA1 in the Apo-form | | Descriptor: | Calcium uptake protein 1, mitochondrial | | Authors: | Wang, L, Yang, X, Li, S, Shen, Y. | | Deposit date: | 2013-11-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake.
Embo J., 33, 2014
|
|
4NSD
 
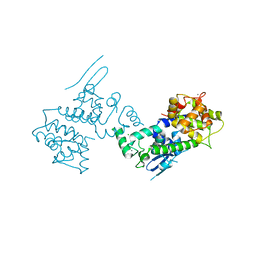 | | Crystal Structure of CBARA1 in the Ca2+ Binding Form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, L, Yang, X, Li, S, Shen, Y. | | Deposit date: | 2013-11-28 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake.
Embo J., 33, 2014
|
|
7U4T
 
 | | Human V-ATPase in state 2 with SidK and mEAK-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MTOR-associated protein MEAK7, ... | | Authors: | Wang, L, Fu, T.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-08-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Identification of mEAK-7 as a human V-ATPase regulator via cryo-EM data mining.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UPT
 
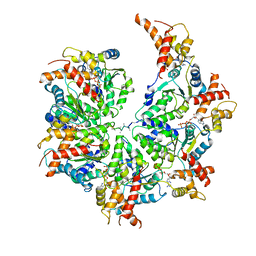 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (open conformation) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
7UPR
 
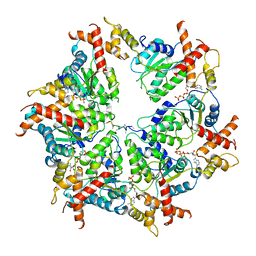 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (closed conformation) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Outer mitochondrial transmembrane helix translocase, ... | | Authors: | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | Deposit date: | 2022-04-16 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
4JR6
 
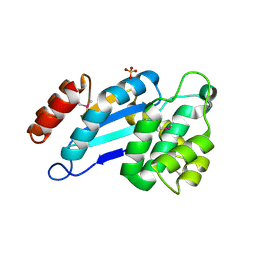 | | Crystal structure of DsbA from Mycobacterium tuberculosis (reduced) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
4JR4
 
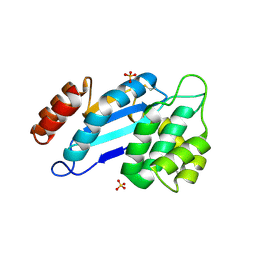 | | Crystal structure of Mtb DsbA (Oxidized) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
6D26
 
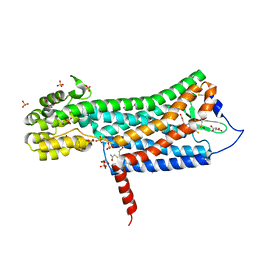 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6D27
 
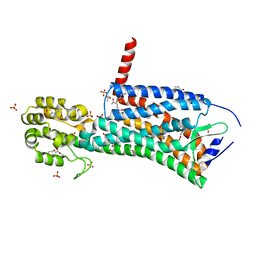 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6EGA
 
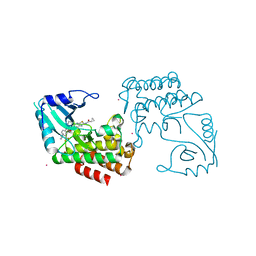 | | IRAK4 in complex with a type II inhibitor | | Descriptor: | 3-{2-[(cyclopropanecarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}-N-{4-[(piperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide, COBALT (II) ION, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, L, Wu, H. | | Deposit date: | 2018-08-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).
J.Biol.Chem., 294, 2019
|
|
6EG9
 
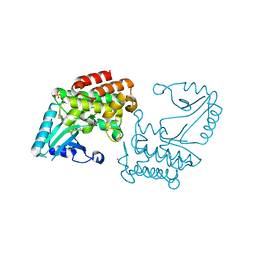 | | IRAK4 in complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, COBALT (II) ION, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, L, Wu, H. | | Deposit date: | 2018-08-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).
J.Biol.Chem., 294, 2019
|
|
6FXD
 
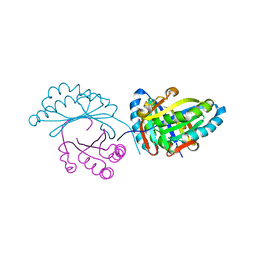 | | Crystal structure of MupZ from Pseudomonas fluorescens | | Descriptor: | MupZ | | Authors: | Wang, L, Parnell, A, Williams, C, Bakar, N.A, van der Kamp, M.W, Simpson, T.J, Race, P.R, Crump, M.P, Willis, C.L. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis
Nat Catal, 2018
|
|
2JRT
 
 | | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides. Northeast Structural Genomics target RhR5 | | Descriptor: | Uncharacterized protein | | Authors: | Wang, L, Chen, C, Nwosu, C, Cunningham, K, Owens, L, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-08-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the protein coded by gene RHOS4_12090 of Rhodobacter sphaeroides.
To be Published
|
|
7E1A
 
 | |
7DV5
 
 | |
