1R0E
 
 | | Glycogen synthase kinase-3 beta in complex with 3-indolyl-4-arylmaleimide inhibitor | | Descriptor: | 3-[3-(2,3-DIHYDROXY-PROPYLAMINO)-PHENYL]-4-(5-FLUORO-1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, CITRATE ANION, Glycogen synthase kinase-3 beta | | Authors: | Allard, J, Nikolcheva, T, Gong, L, Wang, J, Dunten, P, Avnur, Z, Waters, R, Sun, Q, Skinner, B. | | Deposit date: | 2003-09-20 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From genetics to therapeutics: the Wnt pathway and osteoporosis
To be Published
|
|
5MGV
 
 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
3KD7
 
 | |
9CBL
 
 | | Cryo-EM structure of epinephrine-bound alpha-2A-adrenergic receptor in complex with heterotrimeric Gi-protein | | Descriptor: | Endolysin,Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lou, J.S, Su, M, Wang, J, Do, H.N, Miao, Y, Huang, X.Y. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct binding conformations of epinephrine with alpha- and beta-adrenergic receptors.
Exp.Mol.Med., 2024
|
|
9CBM
 
 | | Cryo-EM structure of dexmedetomidine-bound alpha-2A-adrenergic receptor in complex with heterotrimeric Gi-protein | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Endolysin,Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Lou, J.S, Su, M, Wang, J, Do, H.N, Miao, Y, Huang, X.Y. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct binding conformations of epinephrine with alpha- and beta-adrenergic receptors.
Exp.Mol.Med., 2024
|
|
3S0Z
 
 | | Crystal structure of New Delhi Metallo-beta-lactamase (NDM-1) | | Descriptor: | Metallo-beta-lactamase, ZINC ION | | Authors: | Guo, Y, Wang, J, Niu, G.J, Shui, W.Q, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-05-13 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural view of the antibiotic degradation enzyme NDM-1 from a superbug.
Protein Cell, 2011
|
|
3S7X
 
 | | Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Major capsid protein VP1, ... | | Authors: | Neu, U, Wang, J, Stehle, T. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the major capsid proteins of the human KI and WU polyomaviruses.
J.Virol., 85, 2011
|
|
5FKP
 
 | | Crystal structure of the mouse CD1d in complex with the p99 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN-PRESENTING GLYCOPROTEIN CD1D1, BETA 2 MICROGLOBULIN, ... | | Authors: | Girardi, E, Wang, J, Zajonc, D.M. | | Deposit date: | 2015-10-18 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Alpha-Helical Peptide and Lipopeptide Bound to the Non-Classical Mhc Class I Molecule Cd1D
J.Biol.Chem., 291, 2016
|
|
4M6F
 
 | |
6WB8
 
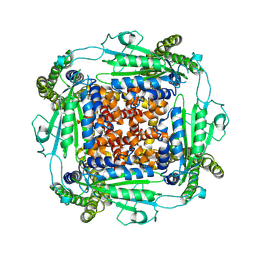 | | Cryo-EM structure of PKD2 C331S disease variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Cao, E, Wang, J, Decaen, P.G. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Molecular dysregulation of ciliary polycystin-2 channels caused by variants in the TOP domain.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WAY
 
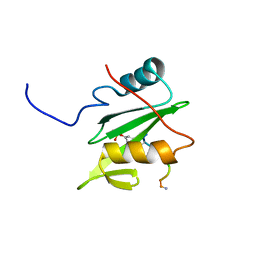 | | C-terminal SH2 domain of p120RasGAP in complex with p190RhoGAP phosphotyrosine peptide | | Descriptor: | Ras GTPase-activating protein 1, Rho GTPase-activating protein 35 | | Authors: | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
4XZC
 
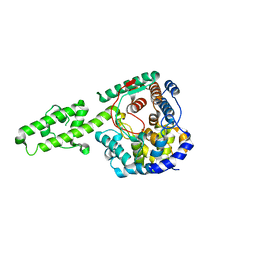 | | The crystal structure of Kupe virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZE
 
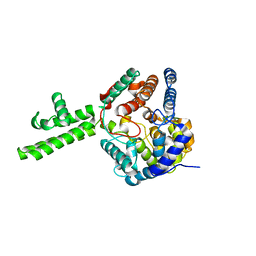 | | The crystal structure of Hazara virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZA
 
 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZ8
 
 | | The crystal structure of Erve virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
6IFZ
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR2-ssDNA complex | | Descriptor: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFR
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-NTR, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Type III-A CRISPR-associated RAMP protein Csm3, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFU
 
 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | Descriptor: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFK
 
 | | Cryo-EM structure of type III-A Csm-CTR1 complex, AMPPNP bound | | Descriptor: | CTR1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | Descriptor: | FAB Heavy chain, FAB Light chain | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6IFN
 
 | | Crystal structure of Type III-A CRISPR Csm complex | | Descriptor: | MANGANESE (II) ION, RNA (32-MER), Type III-A CRISPR-associated RAMP protein Csm3, ... | | Authors: | You, L, Wang, J, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
7RZC
 
 | | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor | | Descriptor: | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor
To be Published
|
|
7SDR
 
 | | Papain-Like Protease of SARS CoV-2 in Complex with Jun9-72-2 Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({methyl[(1R)-1-(naphthalen-1-yl)ethyl]amino}methyl)phenol, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in Complex with Jun9-72-2 Inhibitor
To be Published
|
|
2KRL
 
 | | The ensemble of the solution global structures of the 102-nt ribosome binding structure element of the turnip crinkle virus 3' UTR RNA | | Descriptor: | RNA (102-MER) | | Authors: | Zuo, X, Wang, J, Yu, P, Eyler, D, Xu, H, Starich, M, Tiede, D, Simon, A, Kasprzak, W, Schwieters, C, Shapiro, B. | | Deposit date: | 2009-12-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
