2KTQ
 
 | | OPEN TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3'), DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Li, Y, Waksman, G. | | Deposit date: | 1998-07-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation.
EMBO J., 17, 1998
|
|
1EKB
 
 | | THE SERINE PROTEASE DOMAIN OF ENTEROPEPTIDASE BOUND TO INHIBITOR VAL-ASP-ASP-ASP-ASP-LYS-CHLOROMETHANE | | Descriptor: | ENTEROPEPTIDASE, VAL-ASP-ASP-ASP-ASP-LYK PEPTIDE, ZINC ION | | Authors: | Fuetterer, K, Lu, D, Sadler, J.E, Waksman, G. | | Deposit date: | 1999-05-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enteropeptidase light chain complexed with an analog of the trypsinogen activation peptide.
J.Mol.Biol., 292, 1999
|
|
3CRE
 
 | |
1EYG
 
 | |
3CRF
 
 | |
3CL3
 
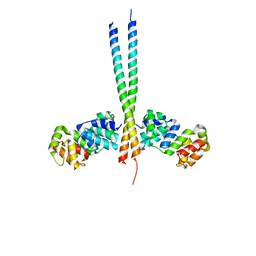 | | Crystal Structure of a vFLIP-IKKgamma complex: Insights into viral activation of the IKK signalosome | | Descriptor: | NF-kappa-B essential modulator, ORF K13 | | Authors: | Bagneris, C, Ageichik, A.V, Cronin, N, Boshoff, C, Waksman, G, Barrett, T. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a vFlip-IKKgamma complex: insights into viral activation of the IKK signalosome.
Mol.Cell, 30, 2008
|
|
1G6O
 
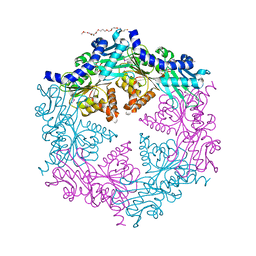 | | CRYSTAL STRUCTURE OF THE HELICOBACTER PYLORI ATPASE, HP0525, IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAG-ALPHA, DI(HYDROXYETHYL)ETHER | | Authors: | Yeo, H.J, Savvides, S.N, Herr, A.B, Lanka, E, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-07 | | Release date: | 2001-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system.
Mol.Cell, 6, 2000
|
|
1IS0
 
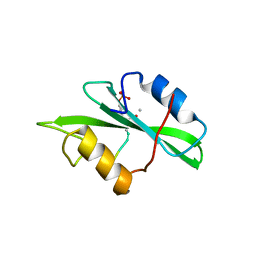 | | Crystal Structure of a Complex of the Src SH2 Domain with Conformationally Constrained Peptide Inhibitor | | Descriptor: | AY0 GLU GLU ILE peptide, Tyrosine-protein kinase transforming protein SRC | | Authors: | Davidson, J.P, Lubman, O, Rose, T, Waksman, G, Martin, S.F. | | Deposit date: | 2001-11-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calorimetric and structural studies of 1,2,3-trisubstituted cyclopropanes as conformationally constrained peptide inhibitors of Src SH2 domain binding.
J.Am.Chem.Soc., 124, 2002
|
|
5KTQ
 
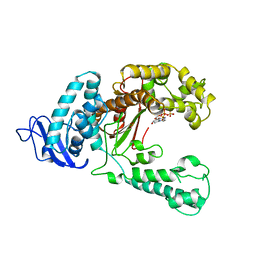 | | LARGE FRAGMENT OF TAQ DNA POLYMERASE BOUND TO DCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, PROTEIN (DNA POLYMERASE I) | | Authors: | Li, Y, Kong, Y, Korolev, S, Waksman, G. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the Klenow fragment of Thermus aquaticus DNA polymerase I complexed with deoxyribonucleoside triphosphates.
Protein Sci., 7, 1998
|
|
5LER
 
 | | Structure of the bacterial sex F pilus (13.2 Angstrom rise) | | Descriptor: | Pilin, [(2~{S})-3-[[(2~{R})-2,3-bis(oxidanyl)propoxy]-oxidanyl-phosphoryl]oxy-2-hexadec-9-enoyloxy-propyl] hexadecanoate | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-30 | | Release date: | 2016-11-02 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
5LEG
 
 | | Structure of the bacterial sex F pilus (pED208) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, Pilin | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-29 | | Release date: | 2016-09-28 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
5LFB
 
 | | Structure of the bacterial sex F pilus (12.5 Angstrom rise) | | Descriptor: | Pilin, [(2~{S})-3-[[(2~{R})-2,3-bis(oxidanyl)propoxy]-oxidanyl-phosphoryl]oxy-2-hexadec-9-enoyloxy-propyl] hexadecanoate | | Authors: | Costa, T.R.D, Ilangovan, I, Ukleja, M, Redzej, A, Santini, J.M, Smith, T.K, Egelman, E.H, Waksman, G. | | Deposit date: | 2016-06-30 | | Release date: | 2016-11-02 | | Last modified: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex.
Cell, 166, 2016
|
|
6EXE
 
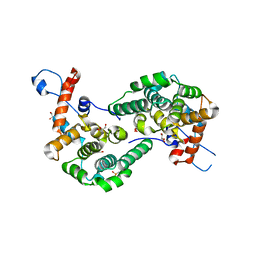 | |
6EXC
 
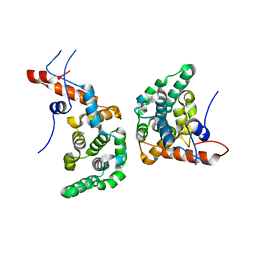 | |
6EXA
 
 | |
6EXD
 
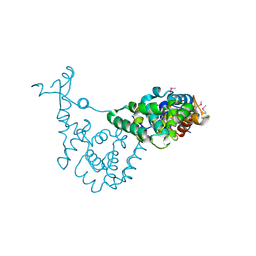 | |
6EXB
 
 | |
6GEB
 
 | |
6GEF
 
 | |
5N6X
 
 | | Crystal structure of the Legionella effector WipA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-MERCAPTOETHANOL, BROMIDE ION, ... | | Authors: | Pinotsis, N, Waksman, G. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-19 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the WipA protein reveals a novel tyrosine protein phosphatase effector from Legionella pneumophila.
J. Biol. Chem., 292, 2017
|
|
5N72
 
 | |
6GYB
 
 | | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri | | Descriptor: | VirB10 protein, VirB7, VirB9 protein | | Authors: | Sgro, G.G, Costa, T.R.D, Farah, C.S, Waksman, G. | | Deposit date: | 2018-06-28 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri.
Nat Microbiol, 3, 2018
|
|
1SRU
 
 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
1THP
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225P MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, PROTEIN (THROMBIN HEAVY CHAIN), PROTEIN (THROMBIN LIGHT CHAIN) | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1TQ0
 
 | | Crystal structure of the potent anticoagulant thrombin mutant W215A/E217A in free form | | Descriptor: | Prothrombin | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
