4U8T
 
 | |
1NY1
 
 | | CRYSTAL STRUCTURE OF B. SUBTILIS POLYSACCHARIDE DEACETYLASE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR127. | | Descriptor: | Probable polysaccharide deacetylase pdaA | | Authors: | Forouhar, F, Edstrom, W, Khan, J, Ma, L, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-11 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Polysaccharide Deacetylase (PDAA_BACSU) from B. Subtilis (Pdaa_Bacsu) Northeast Structural Genomics Research Consortium (Nesg) Target Sr127
To be Published
|
|
3SWV
 
 | | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A | | Descriptor: | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-14 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BrefeldinA-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A
To be Published
|
|
3SY1
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, UPF0001 protein yggS | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Maglaqui, M, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.465 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR70
To be Published
|
|
3SXW
 
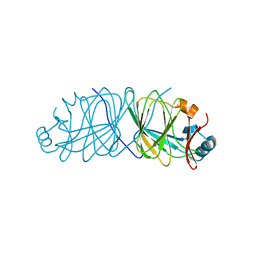 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69. | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Patel, P, Xiao, R, Maglaqui, M, Ciccosanti, C, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69.
To be Published
|
|
3TC6
 
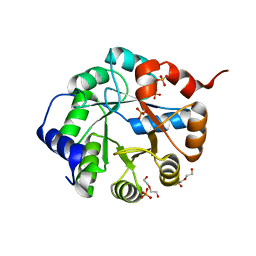 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3TC7
 
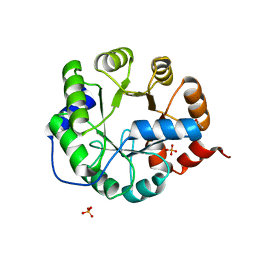 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3E23
 
 | | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299 | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, uncharacterized protein RPA2492 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-05 | | Release date: | 2008-09-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299
To be Published
|
|
3E5Z
 
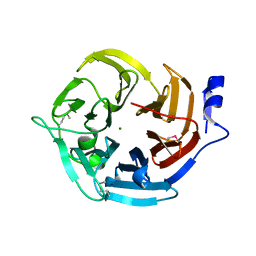 | | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130. | | Descriptor: | MAGNESIUM ION, putative Gluconolactonase | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Tong, S.N, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | X-Ray structure of the putative gluconolactonase in protein family PF08450. Northeast Structural Genomics Consortium target DrR130.
To be Published
|
|
1SBK
 
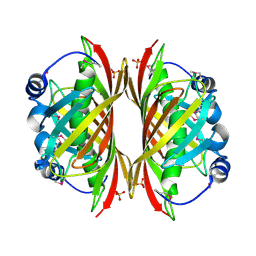 | | X-RAY STRUCTURE OF YDII_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER29. | | Descriptor: | Hypothetical protein ydiI, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Lee, I, Forouhar, F, Ma, L, Chiang, Y, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Structure of YDII_ECOLI Northeast Structural Genomics Consortium Target ER29
To be Published
|
|
5BTE
 
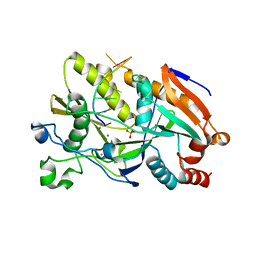 | |
5BTH
 
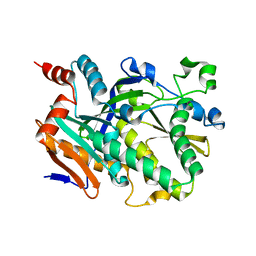 | | Crystal structure of Candida albicans Rai1 | | Descriptor: | Decapping nuclease RAI1 | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5BUD
 
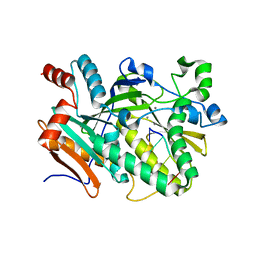 | |
5BTO
 
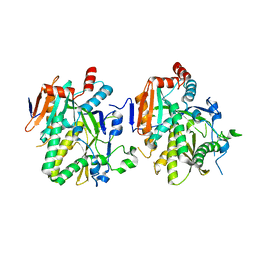 | |
5BTB
 
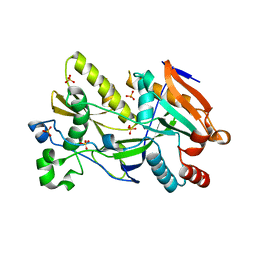 | | Crystal Structure of Ashbya gossypii Rai1 | | Descriptor: | AFR263Cp, SULFATE ION | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5CS4
 
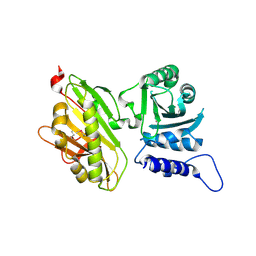 | |
5CS0
 
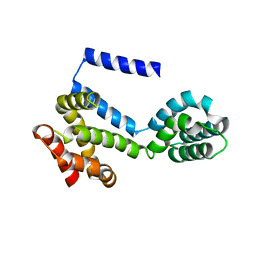 | |
5CSK
 
 | |
5CSA
 
 | |
5CSL
 
 | |
7K95
 
 | | Crystal structure of human CPSF30 in complex with hFip1 | | Descriptor: | Isoform 2 of Cleavage and polyadenylation specificity factor subunit 4, Pre-mRNA 3'-end-processing factor FIP1, ZINC ION | | Authors: | Hamilton, K, Tong, L. | | Deposit date: | 2020-09-28 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism for the interaction between human CPSF30 and hFip1.
Genes Dev., 34, 2020
|
|
7KZ8
 
 | |
7KZ9
 
 | | Crystal structure of Pseudomonas sp. PDC86 substrate-binding protein Aapf in complex with a signaling molecule HEHEAA | | Descriptor: | GLYCEROL, N,N~2~-bis(2-hydroxyethyl)glycinamide, Peptide/nickel transport system substrate-binding protein AapF, ... | | Authors: | Luo, S, Dadhwal, P, Tong, L. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a bacterial Pip system plant effector recognition protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2OKA
 
 | | Crystal structure of Q9HYQ7_PSEAE from Pseudomonas aeruginosa. Northeast Structural Genomics Consortium target PaR82 | | Descriptor: | Hypothetical protein | | Authors: | Benach, J, Neely, H, Seetharaman, J, Chen, X.C, Fang, Y, Cunningham, K, Owens, L, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-01-16 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Q9HYQ7_PSEAE from Pseudomonas aeruginosa
To be Published
|
|
2P0G
 
 | | Crystal structure of Selenoprotein W-related protein from Vibrio cholerae. Northeast Structural Genomics target VcR75 | | Descriptor: | Selenoprotein W-related protein | | Authors: | Benach, J, Neely, H, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Selenoprotein W-related protein from Vibrio cholerae.
To be Published
|
|
