7C9W
 
 | | E30 F-particle in complex with CD55 | | 分子名称: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | 著者 | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | 登録日 | 2020-06-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9U
 
 | | Echovirus 30 E-particle | | 分子名称: | VP0, VP1, VP3 | | 著者 | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | 登録日 | 2020-06-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7CRB
 
 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | 分子名称: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | 著者 | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | 登録日 | 2020-08-13 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7C9T
 
 | | Echovirus 30 A-particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | 登録日 | 2020-06-07 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
5XMM
 
 | | FLA-E*01801-167W/S | | 分子名称: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | 著者 | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | 登録日 | 2017-05-15 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
5XMF
 
 | | Crystal structure of feline MHC class I for 2,1 angstrom | | 分子名称: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | 著者 | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | 登録日 | 2017-05-15 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
6KY8
 
 | | Crystal structure of ASFV dUTPase | | 分子名称: | E165R | | 著者 | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | 登録日 | 2019-09-17 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
6KY9
 
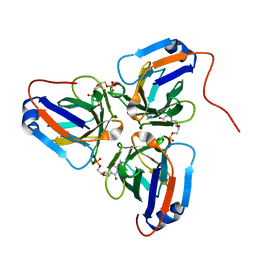 | | Crystal structure of ASFV dUTPase and UMP complex | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | 著者 | Li, C, Chai, Y, Song, H, Qi, J, Sun, Y, Gao, G.F. | | 登録日 | 2019-09-17 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target.
Mbio, 10, 2019
|
|
7XDD
 
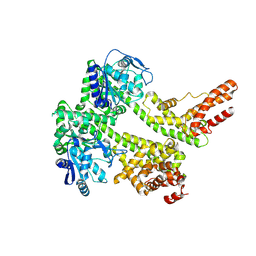 | | Cryo-EM structure of EDS1 and PAD4 | | 分子名称: | Lipase-like PAD4, Protein EDS1 | | 著者 | Huang, S.J, Jia, A.L, Sun, Y, Han, Z.F, Chai, J.J. | | 登録日 | 2022-03-26 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-10 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
6AKS
 
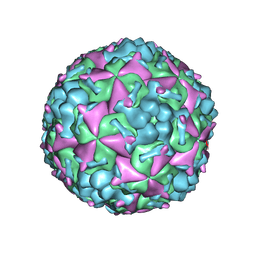 | | Cryo-EM structure of CVA10 mature virus | | 分子名称: | SPHINGOSINE, VP1, VP2, ... | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKT
 
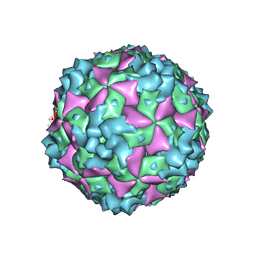 | | Cryo-EM structure of CVA10 A-particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKU
 
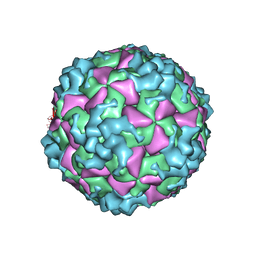 | | Cryo-EM structure of CVA10 empty particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-09-03 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
7F0D
 
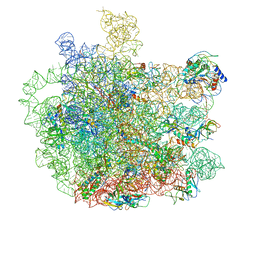 | | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosome subunit bound with clarithromycin | | 分子名称: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Zhang, W, Sun, Y, Gao, N, Li, Z. | | 登録日 | 2021-06-03 | | 公開日 | 2022-06-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium tuberculosis 50S ribosomal subunit bound with clarithromycin reveals dynamic and specific interactions with macrolides.
Emerg Microbes Infect, 11, 2022
|
|
7FIP
 
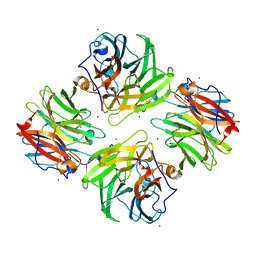 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | 分子名称: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | 著者 | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | 分子名称: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | 著者 | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIS
 
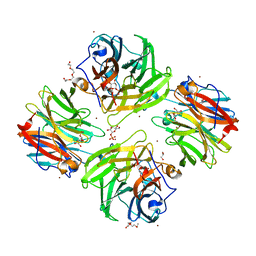 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with mannose 1-phosphate (M1P) | | 分子名称: | 1-O-phosphono-alpha-D-mannopyranose, Beta-1,2-mannobiose phosphorylase, GLYCEROL, ... | | 著者 | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIR
 
 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with 1,4-mannobiose | | 分子名称: | Beta-1,2-mannobiose phosphorylase, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | 著者 | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7ZBT
 
 | | Subtomogram averaging of Rubisco from native Halothiobacillus carboxysomes | | 分子名称: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small subunit | | 著者 | Ni, T, Zhu, Y, Yu, X, Sun, Y, Liu, L, Zhang, P. | | 登録日 | 2022-03-24 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-01-18 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and assembly of cargo Rubisco in two native alpha-carboxysomes.
Nat Commun, 13, 2022
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | 分子名称: | FAB Heavy chain, FAB Light chain | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.907 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X45
 
 | | Crystal structure of 2A protease from Human rhinovirus C15 | | 分子名称: | ZINC ION, protease 2A | | 著者 | Ling, H, Yang, P, Shaw, N, Sun, Y, Wang, X. | | 登録日 | 2017-02-10 | | 公開日 | 2018-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | Structural view of the 2A protease from human rhinovirus C15.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
6MS7
 
 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | 分子名称: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | 著者 | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | 登録日 | 2018-10-16 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
7DFV
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | 分子名称: | NAD+ hydrolase (NADase) | | 著者 | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | 登録日 | 2020-11-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7XC2
 
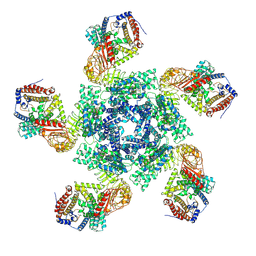 | | Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Avirulence factor, CNL9 | | 著者 | Alexander, F, Li, E.T, Aaron, L, Deng, Y.N, Sun, Y, Chai, J.J. | | 登録日 | 2022-03-22 | | 公開日 | 2022-09-21 | | 最終更新日 | 2022-11-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A wheat resistosome defines common principles of immune receptor channels.
Nature, 610, 2022
|
|
7BX7
 
 | |
