1MU2
 
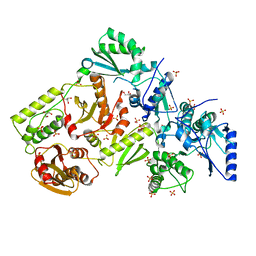 | | CRYSTAL STRUCTURE OF HIV-2 REVERSE TRANSCRIPTASE | | 分子名称: | GLYCEROL, HIV-2 RT, SULFATE ION | | 著者 | Ren, J, Bird, L.E, Chamberlain, P.P, Stewart-Jones, G.B, Stuart, D.I, Stammers, D.K. | | 登録日 | 2002-09-23 | | 公開日 | 2002-10-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8R1C
 
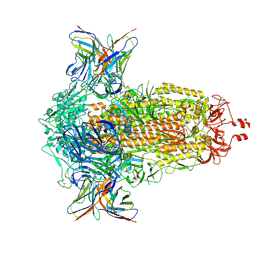 | | SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-2 fab heavy chain, SD1-2 fab light chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-01 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R1D
 
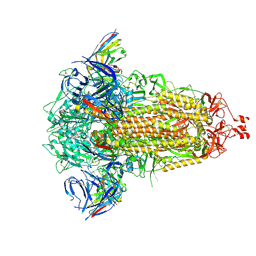 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-01 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-04-10 | | 実験手法 | ELECTRON MICROSCOPY (2.37 Å) | | 主引用文献 | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R8K
 
 | |
2X7L
 
 | | Implications of the HIV-1 Rev dimer structure at 3.2A resolution for multimeric binding to the Rev response element | | 分子名称: | FAB HEAVY CHAIN, FAB LIGHT CHAIN, PROTEIN REV | | 著者 | DiMattia, M.A, Watts, N.R, Stahl, S.J, Rader, C, Wingfield, P.T, Stuart, D.I, Steven, A.C, Grimes, J.M. | | 登録日 | 2010-03-01 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Implications of the HIV-1 Rev Dimer Structure at 3. 2 A Resolution for Multimeric Binding to the Rev Response Element.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1AHC
 
 | |
1AHB
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | 分子名称: | ALPHA-MOMORCHARIN, FORMYCIN-5'-MONOPHOSPHATE | | 著者 | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | 登録日 | 1994-01-07 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1AHA
 
 | | THE N-GLYCOSIDASE MECHANISM OF RIBOSOME-INACTIVATING PROTEINS IMPLIED BY CRYSTAL STRUCTURES OF ALPHA-MOMORCHARIN | | 分子名称: | ADENINE, ALPHA-MOMORCHARIN | | 著者 | Ren, J, Wang, Y, Dong, Y, Stuart, D.I. | | 登録日 | 1994-01-07 | | 公開日 | 1994-06-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The N-glycosidase mechanism of ribosome-inactivating proteins implied by crystal structures of alpha-momorcharin.
Structure, 2, 1994
|
|
1B8M
 
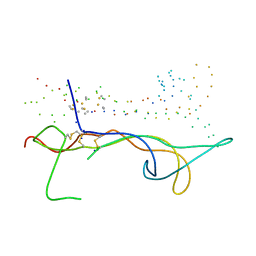 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | 分子名称: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | 著者 | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | 登録日 | 1999-02-01 | | 公開日 | 1999-02-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B98
 
 | | NEUROTROPHIN 4 (HOMODIMER) | | 分子名称: | CHLORIDE ION, PROTEIN (NEUROTROPHIN-4) | | 著者 | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | 登録日 | 1999-02-22 | | 公開日 | 1999-02-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
2BTV
 
 | | ATOMIC MODEL FOR BLUETONGUE VIRUS (BTV) CORE | | 分子名称: | PROTEIN (VP3 CORE PROTEIN), PROTEIN (VP7 CORE PROTEIN) | | 著者 | Grimes, J.M, Burroughs, J.N, Gouet, P, Diprose, J.M, Malby, R, Zientras, S, Mertens, P.P.C, Stuart, D.I. | | 登録日 | 1998-09-05 | | 公開日 | 1998-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | The atomic structure of the bluetongue virus core.
Nature, 395, 1998
|
|
2BRY
 
 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | 登録日 | 2005-05-13 | | 公開日 | 2005-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2C4C
 
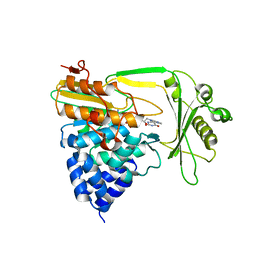 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | 著者 | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | 登録日 | 2005-10-18 | | 公開日 | 2005-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2CME
 
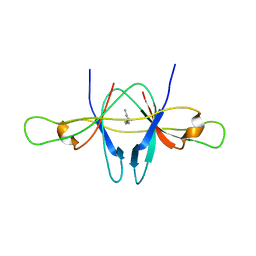 | | The crystal structure of SARS coronavirus ORF-9b protein | | 分子名称: | DECANE, HYPOTHETICAL PROTEIN 5 | | 著者 | Meier, C, Aricescu, A.R, Assenberg, R, Aplin, R.T, Gilbert, R.J.C, Grimes, J.M, Stuart, D.I. | | 登録日 | 2006-05-06 | | 公開日 | 2006-07-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The Crystal Structure of Orf-9B, a Lipid Binding Protein from the Sars Coronavirus.
Structure, 14, 2006
|
|
7PS6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-44 and Beta-54 Fabs | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-44 Fab heavy chain, Beta-44 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-09-22 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
2CDG
 
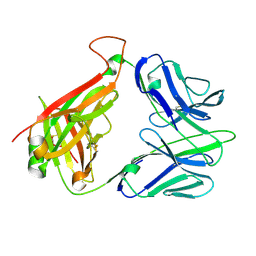 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) | | 分子名称: | TCR 5E | | 著者 | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | 登録日 | 2006-01-23 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
7Q0G
 
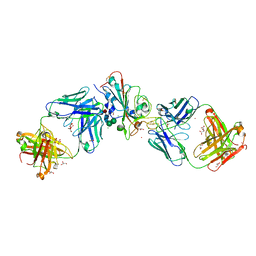 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-49 and FI-3A Fabs | | 分子名称: | Beta-49 Fab heavy chain, Beta-49 Fab light chain, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0H
 
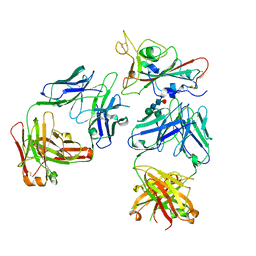 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-50 and Beta-54 | | 分子名称: | Beta-50 Fab heavy chain, Beta-50 Fab light chain, Beta-54 Fab heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0I
 
 | | Crystal structure of the N-terminal domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-43 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
2CDE
 
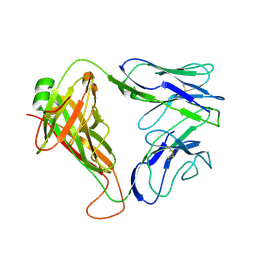 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | 分子名称: | INKT-TCR | | 著者 | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | 登録日 | 2006-01-23 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2CDF
 
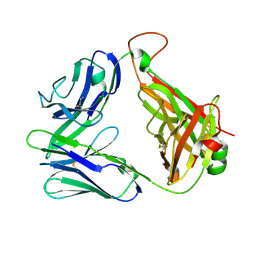 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5E) | | 分子名称: | TCR 5E | | 著者 | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | 登録日 | 2006-01-23 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
7Q0A
 
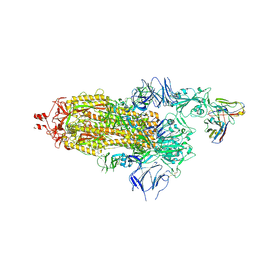 | | SARS-CoV-2 Spike ectodomain with Fab FI3A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FI3A fab Light chain, ... | | 著者 | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | 登録日 | 2021-10-14 | | 公開日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structures and therapeutic potential of anti-RBD human monoclonal antibodies against SARS-CoV-2.
Theranostics, 12, 2022
|
|
1GPB
 
 | |
2DBE
 
 | | CRYSTAL STRUCTURE OF A BERENIL-DODECANUCLEOTIDE COMPLEX: THE ROLE OF WATER IN SEQUENCE-SPECIFIC LIGAND BINDING | | 分子名称: | BERENIL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Brown, D.G, Sanderson, M.R, Skelly, J.V, Jenkins, T.C, Brown, T, Garman, E, Stuart, D.I, Neidle, S. | | 登録日 | 1990-03-19 | | 公開日 | 1991-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a berenil-dodecanucleotide complex: the role of water in sequence-specific ligand binding.
EMBO J., 9, 1990
|
|
1HHS
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | 分子名称: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | 著者 | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | 登録日 | 2000-12-28 | | 公開日 | 2001-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
