7DAO
 
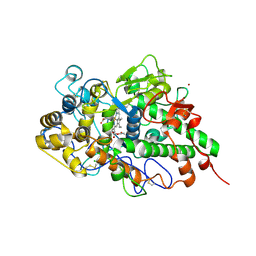 | | Crystal structure of native yak lactoperoxidase at 2.28 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Ahmad, N, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-10-16 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of native yak lactoperoxidase at 2.28 A resolution
To Be Published
|
|
7DMR
 
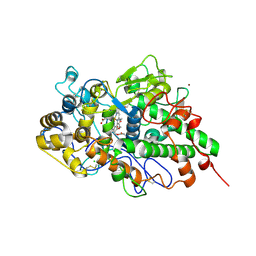 | | Crystal structure of potassium induced heme modification in yak lactoperoxidase at 2.20 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Rani, C, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-12-06 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
5DWF
 
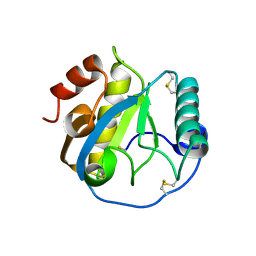 | | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Singh, P.K, Yadav, S.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein, PGRP-S from camel with ethylene glycol at 1.83 A resolution
To Be Published
|
|
8JUK
 
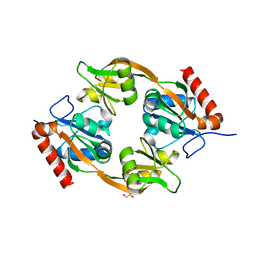 | |
1N76
 
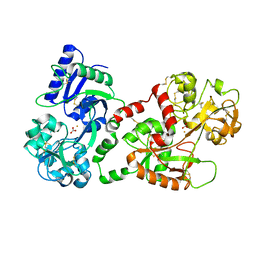 | | CRYSTAL STRUCTURE OF HUMAN SEMINAL LACTOFERRIN AT 3.4 A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN, ... | | Authors: | Kumar, J, Weber, W, Munchau, S, Yadav, S, Singh, S.B, Sarvanan, K, Paramsivam, M, Sharma, S, Kaur, P, Bhushan, A, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of human seminal lactoferrin at 3.4A resolution
Indian J.Biochem.Biophys., 40, 2003
|
|
1OXR
 
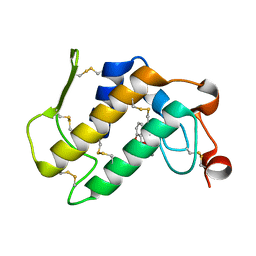 | | Aspirin induces its Anti-inflammatory effects through its specific binding to Phospholipase A2: Crystal structure of the complex formed between Phospholipase A2 and Aspirin at 1.9A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Singh, R.K, Ethayathulla, A.S, Jabeen, T, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2003-04-03 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Aspirin induces its anti-inflammatory effects through its specific binding to phospholipase A2: crystal structure of the complex formed between phospholipase A2 and aspirin at 1.9 angstroms resolution.
J.Drug Target., 13, 2005
|
|
1OYO
 
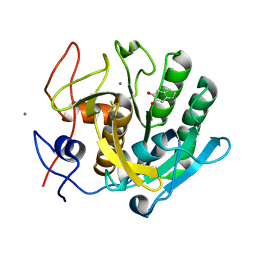 | | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution | | Descriptor: | 3H-INDOLE-5,6-DIOL, CALCIUM ION, Proteinase K | | Authors: | Singh, N, Sharma, S, Kumar, S, Raman, G, Singh, T.P. | | Deposit date: | 2003-04-06 | | Release date: | 2003-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution
To be Published
|
|
1OYF
 
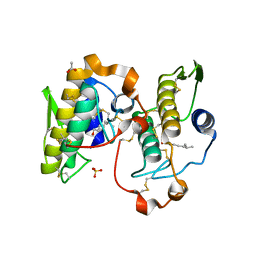 | | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol | | Descriptor: | 6-METHYLHEPTAN-1-OL, ACETIC ACID, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-04-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Russelles viper (Daboia russellii pulchella) phospholipase A2 in a complex with venom 6-methyl heptanol
To be Published
|
|
1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | Deposit date: | 2003-04-02 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
1MH7
 
 | |
1PO8
 
 | | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution. | | Descriptor: | HEPTANOIC ACID, Phospholipase A2, SODIUM ION | | Authors: | Singh, G, Jayasankar, J, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2003-06-14 | | Release date: | 2004-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of a complex formed between krait venom phospholipase A2 and heptanoic acid at 2.7 A resolution.
To be Published
|
|
1Q7A
 
 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
1FB2
 
 | | STRUCTURE OF PHOSPHOLIPASE A2 FROM DABOIA RUSSELLI PULCHELLA AT 1.95 | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Chandra, V, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Regulation of catalytic function by molecular association: structure of phospholipase A2 from Daboia russelli pulchella (DPLA2) at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
8J9T
 
 | | Crystal Structure of GyraseA N-terminal at 2.43A Resolution | | Descriptor: | CARBONATE ION, DNA gyrase subunit A | | Authors: | Salman, M, Sachdeva, E, Das, U, Singh, T.P, Ethayathullah, A.S, Kaur, P. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Crystal Structure of GyraseA N-terminal at 2.43A Resolution
to be published
|
|
5WUY
 
 | | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution | | Descriptor: | Chorismate synthase | | Authors: | Iqbal, N, Chaudhary, A, Shukla, K.P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of chorismate synthase from Acinetobacter baumannii at 2.50A resolution
To Be Published
|
|
1PC8
 
 | | Crystal Structure of a novel form of mistletoe lectin from Himalayan Viscum album L. at 3.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Himalayan mistletoe ribosome-inactivating protein, ... | | Authors: | Mishra, V, Ethayathulla, A.S, Paramasivam, M, Singh, G, Yadav, S, Kaur, P, Sharma, R.S, Babu, C.R, Singh, T.P. | | Deposit date: | 2003-05-16 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a novel ribosome-inactivating protein from a hemi-parasitic plant inhabiting the northwestern Himalayas.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2TPI
 
 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | ISOLEUCINE, MERCURY (II) ION, TRYPSIN INHIBITOR, ... | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
4ZC1
 
 | |
4ZGB
 
 | | Structure of untreated lipase from Thermomyces lanuginosa at 2.3 A resolution | | Descriptor: | Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-04-22 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhancement of stability of a lipase by subjecting to three phase partitioning (TPP): structures of native and TPP-treated lipase from Thermomyces lanuginosa
Sustain Chem Process, 2015
|
|
4ZT8
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a pyrimidine base, cytosine at 1.98 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOPYRIMIDIN-2(1H)-ONE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-14 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
5B6P
 
 | | Structure of the dodecameric type-II dehydrogenate dehydratase from Acinetobacter baumannii at 2.00 A resolution | | Descriptor: | 3-dehydroquinate dehydratase, SULFATE ION | | Authors: | Kumar, M, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding studies and structure determination of the recombinantly produced type-II 3-dehydroquinate dehydratase from Acinetobacter baumannii.
Int. J. Biol. Macromol., 94, 2017
|
|
4ZU0
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine monophosphate at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4ZZ6
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine triphosphate at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4Y55
 
 | | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Gupta, A, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-11 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Buffalo lactoperoxidase with Rhodanide at 2.09 Angstrom resolution
To Be Published
|
|
1I6B
 
 | |
